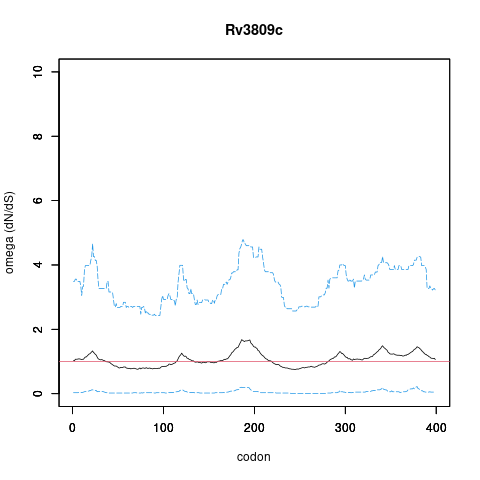
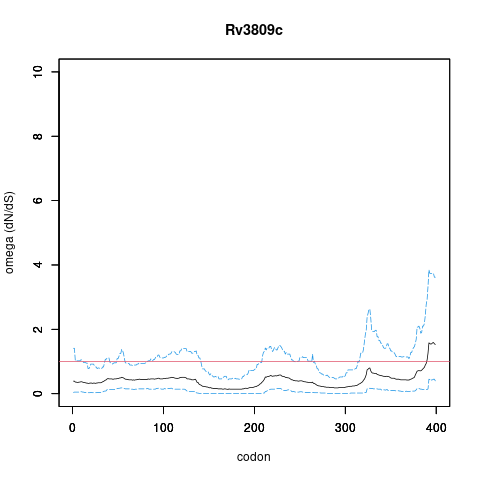
| Property | Value | Creator | Evidence | PMID | Comment |
| Name | UDP-Galactopyranose mutase: Formation of UDP-Galf from UDP-Galp. UDP-Galf is the donor of galactosyl residues | mjackson | IDA | | UDP-Galactofuranose synthesis |
| Citation | Biosynthetic origin of mycobacterial cell wall galactofuranosyl residues. A. Weston,RJ. Stern,RE. Lee,PM. Nassau,D. Monsey,SL. Martin,MS. Scherman,GS. Besra,K. Duncan,MR. McNeil Tuber. Lung Dis. 1997 | mmcneil | | 9692181 | UDP-Galactopyranose Mutase: Formation of UDP-Galf from UDP-Galp. UDP-Galf is the donor of galactosyl residues' enzymatic evidence |
| Term | GO:0009226 nucleotide-sugar biosynthetic process - NR | mmcneil | NR | | UDP-Galactopyranose Mutase: Formation of UDP-Galf from UDP-Galp. UDP-Galf is the donor of galactosyl residues' enzymatic evidence
A. Weston,RJ. Stern,RE. Lee,PM. Nassau,D. Monsey,SL. Martin,MS. Scherman,GS. Besra,K. Duncan,MR. McNeil Biosynthetic origin of mycobacterial cell wall galactofuranosyl residues. Tuber. Lung Dis. 1997 |
| Citation | Galactofuranose biosynthesis in Escherichia coli K-12: identification and cloning of UDP-galactopyranose mutase. authors,PM. Nassau,SL. Martin,RE. Brown,A. Weston,D. Monsey,MR. McNeil,K. Duncan J. Bacteriol. 1996 | jjmcfadden | | 8576037 | Inferred from direct assay |
| Term | EC:5.4.99.9 UDP-galactopyranose mutase. - NR | jjmcfadden | NR | | Inferred from direct assay
authors,PM. Nassau,SL. Martin,RE. Brown,A. Weston,D. Monsey,MR. McNeil,K. Duncan Galactofuranose biosynthesis in Escherichia coli K-12: identification and cloning of UDP-galactopyranose mutase. J. Bacteriol. 1996 |
| Citation | Galactofuranose biosynthesis in Escherichia coli K-12: identification and cloning of UDP-galactopyranose mutase. authors,PM. Nassau,SL. Martin,RE. Brown,A. Weston,D. Monsey,MR. McNeil,K. Duncan J. Bacteriol. 1996 | extern:JZUCKER | | 8576037 | Inferred from direct assay |
| Term | EC:5.4.99.9 UDP-galactopyranose mutase. - NR | extern:JZUCKER | NR | | Inferred from direct assay
authors,PM. Nassau,SL. Martin,RE. Brown,A. Weston,D. Monsey,MR. McNeil,K. Duncan Galactofuranose biosynthesis in Escherichia coli K-12: identification and cloning of UDP-galactopyranose mutase. J. Bacteriol. 1996 |
| Citation | Drug targeting Mycobacterium tuberculosis cell wall synthesis: development of a microtiter plate-based screen for UDP-galactopyranose mutase and identification of an inhibitor from a uridine-based library. authors,MS. Scherman,KA. Winans,RJ. Stern,V. Jones,CR. Bertozzi,MR. McNeil Antimicrob. Agents Chemother. 2003 | extern:JZUCKER | | 12499218 | Assay of protein purified to homogeneity from a heterlogous host |
| Term | EC:5.4.99.9 UDP-galactopyranose mutase. - NR | extern:JZUCKER | NR | | Assay of protein purified to homogeneity from a heterlogous host
authors,MS. Scherman,KA. Winans,RJ. Stern,V. Jones,CR. Bertozzi,MR. McNeil Drug targeting Mycobacterium tuberculosis cell wall synthesis: development of a microtiter plate-based screen for UDP-galactopyranose mutase and identification of an inhibitor from a uridine-based library. Antimicrob. Agents Chemother. 2003 |
| Citation | Biosynthetic origin of mycobacterial cell wall galactofuranosyl residues. A. Weston,RJ. Stern,RE. Lee,PM. Nassau,D. Monsey,SL. Martin,MS. Scherman,GS. Besra,K. Duncan,MR. McNeil Tuber. Lung Dis. 1997 | extern:JZUCKER | | 9692181 | Assay of protein partially-purified from a heterlogous host |
| Term | EC:5.4.99.9 UDP-galactopyranose mutase. - NR | extern:JZUCKER | NR | | Assay of protein partially-purified from a heterlogous host
A. Weston,RJ. Stern,RE. Lee,PM. Nassau,D. Monsey,SL. Martin,MS. Scherman,GS. Besra,K. Duncan,MR. McNeil Biosynthetic origin of mycobacterial cell wall galactofuranosyl residues. Tuber. Lung Dis. 1997 |