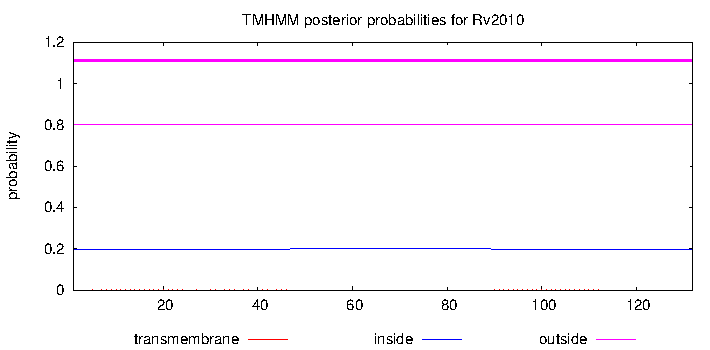
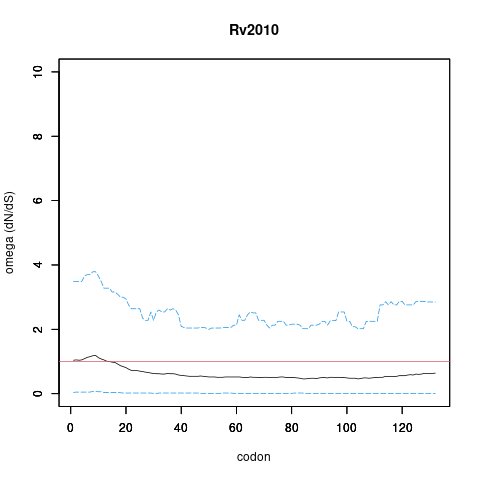
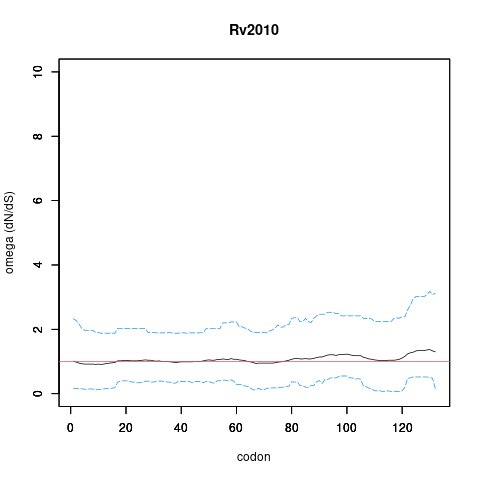
| Property | Value | Creator | Evidence | PMID | Comment |
| Interaction | InhibitedBy Rv2009 | shahanup86 | IEP | | coexpression(Functional Linkage)
authors,HR. Ramage,LE. Connolly,JS. Cox Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009 |
| Interaction | InhibitedBy Rv2009 | shahanup86 | IEP | | coexpression(Functional Linkage)
authors,J. Robson,JL. McKenzie,R. Cursons,GM. Cook,VL. Arcus The vapBC operon from Mycobacterium smegmatis is an autoregulated toxin-antitoxin module that controls growth via inhibition of translation. J. Mol. Biol. 2009 |
| Citation | Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. authors,HR. Ramage,LE. Connolly,JS. Cox PLoS Genet. 2009 | shahanup86 | IEP | 20011113 | coexpression(Functional Linkage) |
| Interaction | InhibitedBy Rv2009 | shahanup86 | IEP | | coexpression(Functional Linkage)
authors,HR. Ramage,LE. Connolly,JS. Cox Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009 |
| Citation | The vapBC operon from Mycobacterium smegmatis is an autoregulated toxin-antitoxin module that controls growth via inhibition of translation. authors,J. Robson,JL. McKenzie,R. Cursons,GM. Cook,VL. Arcus J. Mol. Biol. 2009 | shahanup86 | IEP | 19445953 | coexpression(Functional Linkage) |
| Interaction | InhibitedBy Rv2009 | shahanup86 | IEP | | coexpression(Functional Linkage)
authors,J. Robson,JL. McKenzie,R. Cursons,GM. Cook,VL. Arcus The vapBC operon from Mycobacterium smegmatis is an autoregulated toxin-antitoxin module that controls growth via inhibition of translation. J. Mol. Biol. 2009 |
| Interaction | RegulatedBy Rv0757 | yamir.moreno | IEP | | Microarrays. mRNA levels of regulated element measured and compared between wild-type and trans-element mutation (knockout, over expression etc.) performed by using microarray (or macroarray) experiments..
SB. Walters, E. Dubnau et al. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 2006 |
| Citation | Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. authors,HR. Ramage,LE. Connolly,JS. Cox PLoS Genet. 2009 | jlew | | 20011113 | VapC homolog, PIN domain, Toxic when expressed in Msmeg, rescued by antitoxin (Rv2009) |
| Symbol | VapC | jlew | | |
authors,BA. Ahidjo,D. Kuhnert,JL. McKenzie,EE. Machowski,BG. Gordhan,V. Arcus,GL. Abrahams,V. Mizrahi VapC toxins from Mycobacterium tuberculosis are ribonucleases that differentially inhibit growth and are neutralized by cognate VapB antitoxins. PLoS ONE 2011 |
| Citation | VapC toxins from Mycobacterium tuberculosis are ribonucleases that differentially inhibit growth and are neutralized by cognate VapB antitoxins. authors,BA. Ahidjo,D. Kuhnert,JL. McKenzie,EE. Machowski,BG. Gordhan,V. Arcus,GL. Abrahams,V. Mizrahi PLoS ONE 2011 | jlew | | 21738782 | None |
| Symbol | VapC15 | jlew | | | We report the heterologous toxicity of these TA loci in Escherichia coli and show that only a few of the M. tuberculosis-encoded toxins can inhibit E. coli growth and have a killing effect. This killing effect can be suppressed by coexpression of the cognate antitoxin.
authors,A. Gupta Killing activity and rescue function of genome-wide toxin-antitoxin loci of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 2009 |
| Citation | Killing activity and rescue function of genome-wide toxin-antitoxin loci of Mycobacterium tuberculosis. authors,A. Gupta FEMS Microbiol. Lett. 2009 | jlew | | 19016878 | We report the heterologous toxicity of these TA loci in Escherichia coli and show that only a few of the M. tuberculosis-encoded toxins can inhibit E. coli growth and have a killing effect. This killing effect can be suppressed by coexpression of the cognate antitoxin. |
| Other | start:2258273 | rslayden | | | Conserved hypothetical protein, similar to Rv1561 |
| Other | stop:258671 | rslayden | | | Conserved hypothetical protein, similar to Rv1561 |
| Other | strand:+ | rslayden | | | Conserved hypothetical protein, similar to Rv1561 |
| Other | start:2258273 | rslayden | | | MTCY48.04c, (38.1%identity in 126 aa overlap) |
| Other | stop:258671 | rslayden | | | MTCY48.04c, (38.1%identity in 126 aa overlap) |
| Other | strand:+ | rslayden | | | MTCY48.04c, (38.1%identity in 126 aa overlap) |