| Property | Value | Creator | Evidence | PMID | Comment |
| Term | TBRXN:ACGAMT UDP-N-acetylglucosamine:undecaprenylphosphate N-acetylglucosamine -1-phosphate transferase - ISS | njamshidi | ISS | |10934214 | See PMID: 10934214, Text by Cole et al (Tuberculosis and the tubercle bacillus) chap 18.
K. Mikusov, T. Yagi et al. Biosynthesis of the galactan component of the mycobacterial cell wall. J. Biol. Chem. 2000 |
| Citation | Biosynthesis of the galactan component of the mycobacterial cell wall. K. Mikusov, T. Yagi et al. J. Biol. Chem. 2000 | njamshidi | IPI | |10934214 | See PMID: 10934214, Text by Cole et al (Tuberculosis and the tubercle bacillus) chap 18. |
| Term | TBRXN:ACGAMT UDP-N-acetylglucosamine:undecaprenylphosphate N-acetylglucosamine -1-phosphate transferase - IPI | njamshidi | IPI | |10934214 | See PMID: 10934214, Text by Cole et al (Tuberculosis and the tubercle bacillus) chap 18.
K. Mikusov, T. Yagi et al. Biosynthesis of the galactan component of the mycobacterial cell wall. J. Biol. Chem. 2000 |
| Citation | Biosynthesis of the galactan component of the mycobacterial cell wall. K. Mikusov, T. Yagi et al. J. Biol. Chem. 2000 | njamshidi | ISS | |10934214 | See PMID: 10934214, Text by Cole et al (Tuberculosis and the tubercle bacillus) chap 18. |
| Interaction | RegulatedBy Rv0757 | yamir.moreno | IEP | | Microarrays. mRNA levels of regulated element measured and compared between wild-type and trans-element mutation (knockout, over expression etc.) performed by using microarray (or macroarray) experiments..
SB. Walters, E. Dubnau et al. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 2006 |
| Interaction | RegulatedBy Rv3286c | yamir.moreno | IEP | | Microarrays. mRNA levels of regulated element measured and compared between wild-type and trans-element mutation (knockout, over expression etc.) performed by using microarray (or macroarray) experiments..
EP. Williams, JH. Lee et al. Mycobacterium tuberculosis SigF regulates genes encoding cell wall-associated proteins and directly regulates the transcriptional regulatory gene phoY1. J. Bacteriol. 2007 |
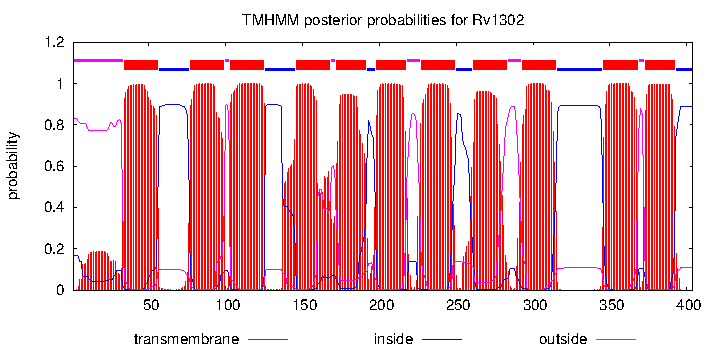
| Symbol | wecA | mjackson | IMP | | GlcNAc-1-phosphate transferase |
| Name | GlcNAc-1-P transferase. Transfers GlcNAc-1-P to decaprenyl phosphate to make GlcNAc-P-P-decaprenyl | mjackson | IMP | | GlcNAc-1-phosphate transferase |
| Symbol | wecA | mmcneil | | | GlcNAc-1-P transferase. Transfers GlcNAc-1-P to decaprenyl phosphate to make GlcNAc-P-P-decaprenyl (complementation of an E. coli LPS mutant)
authors,Y. Jin,Y. Xin,W. Zhang,Y. Ma Mycobacterium tuberculosis Rv1302 and Mycobacterium smegmatis MSMEG_4947 have WecA function and MSMEG_4947 is required for the growth of M. smegmatis. FEMS Microbiol. Lett. 2010 |
| Citation | Mycobacterium tuberculosis Rv1302 and Mycobacterium smegmatis MSMEG_4947 have WecA function and MSMEG_4947 is required for the growth of M. smegmatis. authors,Y. Jin,Y. Xin,W. Zhang,Y. Ma FEMS Microbiol. Lett. 2010 | mmcneil | | 20637039 | GlcNAc-1-P transferase. Transfers GlcNAc-1-P to decaprenyl phosphate to make GlcNAc-P-P-decaprenyl (complementation of an E. coli LPS mutant) |
| Term | GO:0003975 UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase activity - NR | mmcneil | NR | | GlcNAc-1-P transferase. Transfers GlcNAc-1-P to decaprenyl phosphate to make GlcNAc-P-P-decaprenyl (complementation of an E. coli LPS mutant)
authors,Y. Jin,Y. Xin,W. Zhang,Y. Ma Mycobacterium tuberculosis Rv1302 and Mycobacterium smegmatis MSMEG_4947 have WecA function and MSMEG_4947 is required for the growth of M. smegmatis. FEMS Microbiol. Lett. 2010 |
| Citation | Biosynthesis of the linkage region of the mycobacterial cell wall. authors,K. Mikusov,M. Mikus,GS. Besra,I. Hancock,PJ. Brennan J. Biol. Chem. 1996 | extern:JZUCKER | | 8631826 | Assay of unpurified protein expressed in its native host |
| Term | EC:2.4.1.- Transferases. Glycosyltransferases. Hexosyltransferases. - NR | extern:JZUCKER | NR | | Assay of unpurified protein expressed in its native host
authors,K. Mikusov,M. Mikus,GS. Besra,I. Hancock,PJ. Brennan Biosynthesis of the linkage region of the mycobacterial cell wall. J. Biol. Chem. 1996 |