MtbTnDB - interactive tool for exploring a database of published TnSeq datasets for Mtb
TnSeqCorr - genes with correlated TnSeq profiles across ~100 conditions
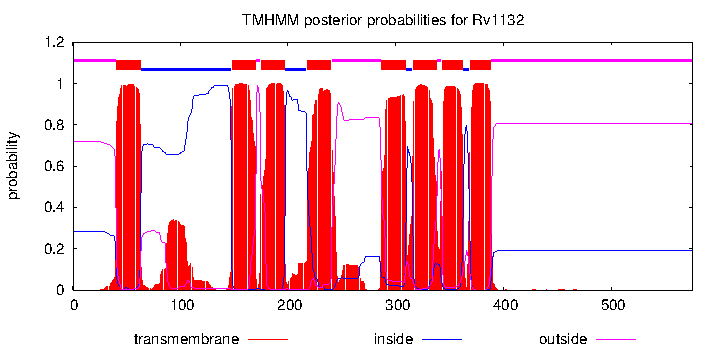
Rv1132/-, gene len: 1730 bp, num TA sites: 27| condition | dataset | call | medium | method | notes |
|---|---|---|---|---|---|
| in-vitro | DeJesus 2017 mBio | non-essential | 7H9 | HMM | fully saturated, 14 TnSeq libraries combined |
| in-vitro | Sassetti 2003 Mol Micro | non-essential | 7H9 | TRASH | essential if hybridization ratio<0.2 |
| in-vivo (mice) | Sassetti 2003 PNAS | non-essential | BL6 mice | TRASH | essential if hybridization ratio<0.4, min over 4 timepoints (1-8 weeks) |
| in-vitro (glycerol) | Griffin 2011 PPath | non-essential | M9 minimal+glycerol | Gumbel | 2 replicates; Padj<0.05 |
| in-vitro (cholesterol) | Griffin 2011 PPath | non-essential | M9 minimal+cholesterol | Gumbel | 3 replicates; Padj<0.05 |
| differentially essential in cholesterol | Griffin 2011 PPath | YES (LFC=-1.18) | cholesterol vs glycerol | resampling-SR | YES if Padj<0.05, else not significant; LFC<0 means less insertions/more essential in cholesterol |
| in-vitro | Smith 2022 eLife | non-essential | 7H9 | HMM | 6 replicates (raw data in Subramaniam 2017, PMID 31752678) |
| in-vivo (mice) | Smith 2022 eLife | non-essential | BL6 mice | HMM | 6 replicates (raw data in Subramaniam 2017, PMID 31752678) |
| differentially essential in mice | Smith 2022 eLife | NO (LFC=0.153) | in-vivo vs in-vitro | ZINB | YES if Padj<0.05, else not significant; LFC<0 means less insertions/more essential in mice |
| in-vitro (minimal) | Minato 2019 mSys | non-essential | minimal medium | HMM | |
| in-vitro (YM rich medium) | Minato 2019 mSys | non-essential | YM rich medium | HMM | 7H9 supplemented with ~20 metabolites (amino acids, cofactors) |
| differentially essential in YM rich medium | Minato 2019 mSys | NO (LFC=-0.48) | YM rich vs minimal medium | resampling | |