Rv0703 (rplW)
Current annotations:
TBCAP: (community-based annotations - see table at bottom of page )
TBDB: 50S ribosomal protein L23
REFSEQ: 50S ribosomal protein L23
PATRIC: LSU ribosomal protein L23p (L23Ae)
TUBERCULIST: 50S ribosomal protein L23 RplW
NCBI: 50S ribosomal protein L23 RplW
updated information (H37Rv4):
gene name: rplW
function:
reference:
Coordinates in H37Rv: 802133 - 802435
Gene length: 303 bp (with stop codon), 100 aa (without stop codon)
Operon:
Trans-membrane region:
Role: II.A.1 - Ribosomal protein synthesis and modification
GO terms:
Reaction(s) (based on iSM810 metabolic model):
Gene Expression Profile Gene Modules Orthologs among selected mycobacteria Protein structure:
Search for Homologs in PDB Top 10 Homologs in PDB (as of Nov 2020): PDB aa ident species PDB title 5V93 100% Mycobacterium tuberculosis Cryo-EM structure of the 70S ribosome from Mycobacterium tuberculosis bound with Capreomycin 5V7Q 100% Mycobacterium tuberculosis Cryo-EM structure of the large ribosomal subunit from Mycobacterium tuberculosis bound with a potent linezolid analog 5ZET 87% Mycobacterium smegmatis str. MC2 155 M. smegmatis P/P state 50S ribosomal subunit 5ZEP 87% Mycobacterium smegmatis str. MC2 155 M. smegmatis hibernating state 70S ribosome structure 5ZEB 87% Mycobacterium smegmatis str. MC2 155 M. Smegmatis P/P state 70S ribosome structure 5XYM 87% Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) Large subunit of Mycobacterium smegmatis 5O61 87% Mycobacterium smegmatis str. MC2 155 The complete structure of the Mycobacterium smegmatis 70S ribosome 5O60 87% Mycobacterium smegmatis str. MC2 155 Structure of the 50S large ribosomal subunit from Mycobacterium smegmatis 6DZP 86% Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) Cryo-EM Structure of Mycobacterium smegmatis C(minus) 50S ribosomal subunit 6DZI 86% Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) Cryo-EM Structure of Mycobacterium smegmatis 70S C(minus) ribosome 70S-MPY complex
Links to additional information on rplW:
Amino Acid Sequence
MATLADPRDIILAPVISEKSYGLLDDNVYTFLVRPDSNKTQIKIAVEKIFAVKVASVNTANRQGKRKRTRTGYGKRKSTKRAIVTLAPGSRPIDLFGAPA
(
Nucleotide sequence available on
KEGG )
Additional Information
MtbTnDB - interactive tool for exploring a database of published TnSeq datasets for Mtb
TnSeqCorr
Rv0703/rplW,
gene len: 302 bp, num TA sites: 5
condition dataset call medium method notes
in-vitro DeJesus 2017 mBio essential 7H9 HMM fully saturated, 14 TnSeq libraries combined
in-vitro Sassetti 2003 Mol Micro essential 7H9 TRASH essential if hybridization ratio<0.2
in-vivo (mice) Sassetti 2003 PNAS no data BL6 mice TRASH essential if hybridization ratio<0.4, min over 4 timepoints (1-8 weeks)
in-vitro (glycerol) Griffin 2011 PPath essential M9 minimal+glycerol Gumbel 2 replicates; Padj<0.05
in-vitro (cholesterol) Griffin 2011 PPath uncertain M9 minimal+cholesterol Gumbel 3 replicates; Padj<0.05
differentially essential in cholesterol Griffin 2011 PPath NO (LFC=0.0) cholesterol vs glycerol resampling-SR YES if Padj<0.05, else not significant; LFC<0 means less insertions/more essential in cholesterol
in-vitro Smith 2022 eLife essential 7H9 HMM 6 replicates (raw data in Subramaniam 2017, PMID 31752678)
in-vivo (mice) Smith 2022 eLife essential BL6 mice HMM 6 replicates (raw data in Subramaniam 2017, PMID 31752678)
differentially essential in mice Smith 2022 eLife NO (LFC=-0.059) in-vivo vs in-vitro ZINB YES if Padj<0.05, else not significant; LFC<0 means less insertions/more essential in mice
in-vitro (minimal) Minato 2019 mSys essential minimal medium HMM
in-vitro (YM rich medium) Minato 2019 mSys growth defect YM rich medium HMM 7H9 supplemented with ~20 metabolites (amino acids, cofactors)
differentially essential in YM rich medium Minato 2019 mSys NO (LFC=0.0) YM rich vs minimal medium resampling
Analysis of Positive Selection in Clinical Isolates
*new*
data from Culviner et al (2025) (55,259 Mtb clinical isolates)
overall pN/pS for Rv0703: 0.634395767
lineage-specific pN/pS in L1: 0.904601372
lineage-specific pN/pS in L2: 0.332302545
lineage-specific pN/pS in L3: 0.352442093
lineage-specific pN/pS in L4: 1.001522948
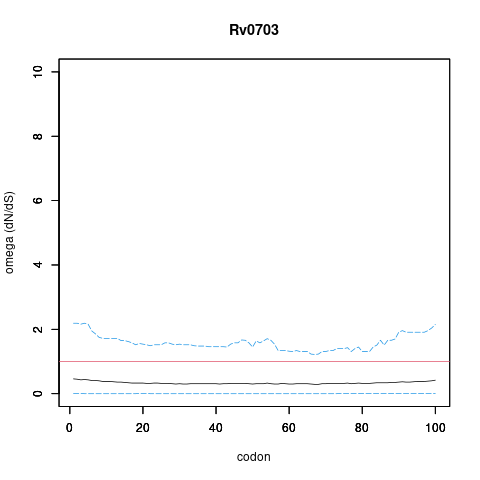
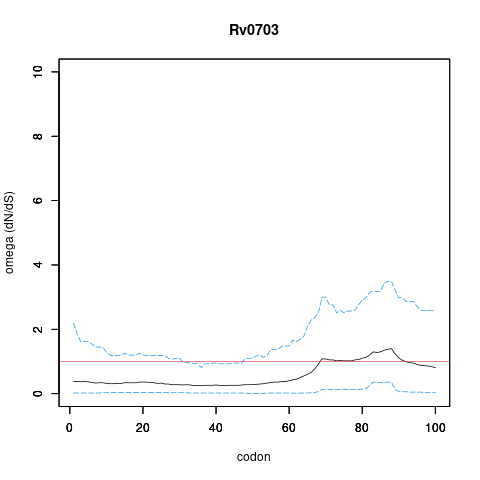
Analysis of dN/dS (omega) in two collections of Mtb clinical isolates using GenomegaMap (Window model) (see description of methods )
Moldova: 2,057 clinical isolates
global set: 5,195 clinical isolates from 15 other countries
In the omega plots, the black line shows the mean estimate of omega (dN/dS) at each codon, and the blue lines are the bounds for the 95% credible interval (95%CI, from MCMC sampling).
A gene is under significant positive selection if the lower-bound of the 95%CI of omega (lower blue line) exceeds 1.0 at any codon.
Moldova (2,057) global set (5,195)
under significant positive selection? NO NO
omega peak height (95%CI lower bound) 0.42 (0.01) 1.4 (0.36)
codons under selection
omega plots
genetic variants* link link
statistics at each codon link link
* example format for variants: "D27 (GAC): D27H (CAC,11)" means "Asp27 (native codon GAC) mutated to His (codon CAC) in 11 isolates"
TnSeq Data No data currently available.
No TnSeq data currently available for this Target.
RNASeq Data No data currently available.
No RNA-Seq data currently available for this Target.
Metabolomic Profiles No data currently available.
No Metabolomic data currently available for this Target.
Proteomic Data No data currently available.
No Proteomic data currently available for this Target.
Regulatory Relationships from Systems Biology
BioCyc
Gene interactions based on ChIPSeq and Transcription Factor Over-Expression (TFOE) (Systems Biology )
NOTE:
Green edges represent the connected genes being classified as differentially essential as a result of the middle gene being knocked out. These interactions are inferred based on RNASeq.
Interactions based on ChIPSeq data
RNA processing and modification
Energy production and conversion
Chromatin structure and dynamics
Amino acid transport and metabolism
Cell cycle control, cell division, chromosome partitioning
Carbohydrate transport and metabolism
Nucleotide transport and metabolism
Lipid transport and metabolism
Coenzyme transport and metabolism
Translation, ribosomal structure and biogenesis
Cell wall/membrane/envelope biogenesis
Replication, recombination and repair
Posttranslational modification, protein turnover, chaperones
Secondary metabolites biosynthesis, transport and catabolism
Inorganic ion transport and metabolism
General function prediction only
Intracellular trafficking, secretion, and vesicular transport
Signal transduction mechanisms
Differentially expressed as result of RNASeq in glycerol environment (Only top 20 genes shown sorted by log fold change with p_adj 0.05).
Conditionally essential as result of TNSeq (Only top 20 genes shown sorted by log fold change with p_adj 0.05).
Binds To:
No bindings to other targets were found.
Bound By:
No bindings from other targets were found.
Binds To:
No bindings to other targets were found.
Bound By:
Upregulates:
Does not upregulate other genes.
Upregulated by:
Not upregulated by other genes.
Downregulates:
Does not downregulate other genes.
Downregulated by:
Not downregulated by other genes.
Property Value Creator Evidence PMID Comment
Interaction PhysicalInteraction Rv0709 ravirajsoni IEP Co-expression (Functional linkage)authors,R. Wang,JT. Prince,EM. Marcotte Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005
Interaction PhysicalInteraction Rv0708 ravirajsoni IEP Co-expression (Functional linkage)authors,R. Wang,JT. Prince,EM. Marcotte Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005
Interaction PhysicalInteraction Rv0707 ravirajsoni IEP Co-expression (Functional linkage)authors,R. Wang,JT. Prince,EM. Marcotte Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005
Interaction PhysicalInteraction Rv0705 ravirajsoni IEP Co-expression (Functional linkage)authors,R. Wang,JT. Prince,EM. Marcotte Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005
Interaction PhysicalInteraction Rv0705 ravirajsoni IEP Co-expression (Functional linkage)authors,R. Wang,JT. Prince,EM. Marcotte Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005
Interaction PhysicalInteraction Rv0706 ravirajsoni IEP Co-expression (Functional linkage)authors,R. Wang,JT. Prince,EM. Marcotte Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005
Interaction PhysicalInteraction Rv0707 ravirajsoni IEP Co-expression (Functional linkage)authors,R. Wang,JT. Prince,EM. Marcotte Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005
Interaction PhysicalInteraction Rv0708 ravirajsoni IEP Co-expression (Functional linkage)authors,R. Wang,JT. Prince,EM. Marcotte Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005
Interaction PhysicalInteraction Rv0709 ravirajsoni IEP Co-expression (Functional linkage)authors,R. Wang,JT. Prince,EM. Marcotte Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005
Interaction PhysicalInteraction Rv0710 ravirajsoni IEP Co-expression (Functional linkage)authors,R. Wang,JT. Prince,EM. Marcotte Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005
Interaction Regulatory Rv0709 ravirajsoni IPI ChIP (Physical interaction)authors,S. Rodrigue,J. Brodeur,PE. Jacques,AL. Gervais,R. Brzezinski,L. Gaudreau Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 2007
Interaction Regulatory Rv0710 ravirajsoni IPI ChIP (Physical interaction)authors,S. Rodrigue,J. Brodeur,PE. Jacques,AL. Gervais,R. Brzezinski,L. Gaudreau Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 2007
Citation Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. authors,R. Wang,JT. Prince,EM. Marcotte Genome Res. 2005 ravirajsoni IEP 16077011 Co-expression (Functional linkage)
Interaction PhysicalInteraction Rv0700 ravirajsoni IEP Co-expression (Functional linkage)authors,R. Wang,JT. Prince,EM. Marcotte Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005
Interaction PhysicalInteraction Rv0701 ravirajsoni IEP Co-expression (Functional linkage)authors,R. Wang,JT. Prince,EM. Marcotte Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005
Interaction PhysicalInteraction Rv0702 ravirajsoni IEP Co-expression (Functional linkage)authors,R. Wang,JT. Prince,EM. Marcotte Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005
Interaction PhysicalInteraction Rv0704 ravirajsoni IEP Co-expression (Functional linkage)authors,R. Wang,JT. Prince,EM. Marcotte Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005
Interaction Regulatory Rv0702 ravirajsoni IPI ChIP (Physical interaction)authors,S. Rodrigue,J. Brodeur,PE. Jacques,AL. Gervais,R. Brzezinski,L. Gaudreau Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 2007
Interaction Regulatory Rv0704 ravirajsoni IPI ChIP (Physical interaction)authors,S. Rodrigue,J. Brodeur,PE. Jacques,AL. Gervais,R. Brzezinski,L. Gaudreau Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 2007
Interaction Regulatory Rv0705 ravirajsoni IPI ChIP (Physical interaction)authors,S. Rodrigue,J. Brodeur,PE. Jacques,AL. Gervais,R. Brzezinski,L. Gaudreau Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 2007
Interaction Regulatory Rv0706 ravirajsoni IPI ChIP (Physical interaction)authors,S. Rodrigue,J. Brodeur,PE. Jacques,AL. Gervais,R. Brzezinski,L. Gaudreau Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 2007
Interaction Regulatory Rv0707 ravirajsoni IPI ChIP (Physical interaction)authors,S. Rodrigue,J. Brodeur,PE. Jacques,AL. Gervais,R. Brzezinski,L. Gaudreau Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 2007
Interaction Regulatory Rv0708 ravirajsoni IPI ChIP (Physical interaction)authors,S. Rodrigue,J. Brodeur,PE. Jacques,AL. Gervais,R. Brzezinski,L. Gaudreau Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 2007
Citation Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. authors,S. Rodrigue,J. Brodeur,PE. Jacques,AL. Gervais,R. Brzezinski,L. Gaudreau J. Bacteriol. 2007 ravirajsoni IPI 17158685 ChIP (Physical interaction)
Interaction Regulatory Rv0700 ravirajsoni IPI ChIP (Physical interaction)authors,S. Rodrigue,J. Brodeur,PE. Jacques,AL. Gervais,R. Brzezinski,L. Gaudreau Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 2007
Interaction Regulatory Rv0701 ravirajsoni IPI ChIP (Physical interaction)authors,S. Rodrigue,J. Brodeur,PE. Jacques,AL. Gervais,R. Brzezinski,L. Gaudreau Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. J. Bacteriol. 2007
Interaction RegulatedBy Rv0348 yamir.moreno IEP Microarrays. mRNA levels of regulated element measured and compared between wild-type and trans-element mutation (knockout, over expression etc.) performed by using microarray (or macroarray) experiments..B. Abomoelak, EA. Hoye et al. mosR, a novel transcriptional regulator of hypoxia and virulence in Mycobacterium tuberculosis. J. Bacteriol. 2009