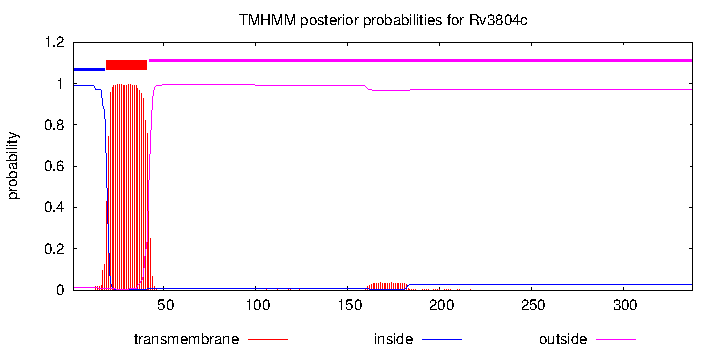
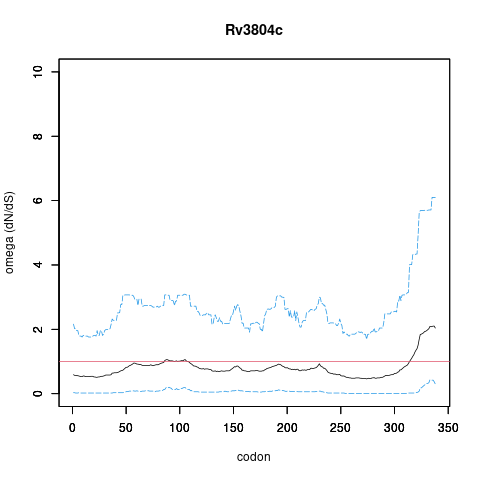
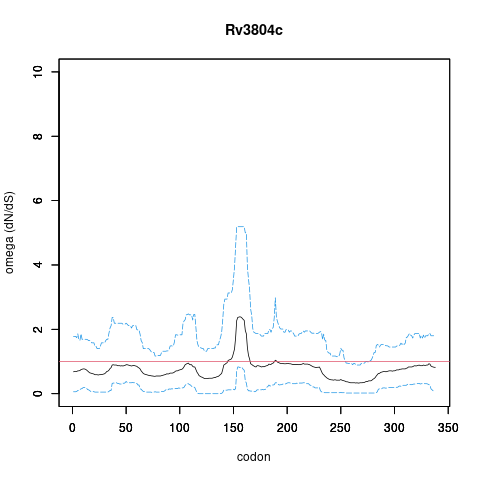
| Property | Value | Creator | Evidence | PMID | Comment |
| Interaction | PhysicalInteraction Rv0129c | prabhakarsmail | IPI | | structural analysis
K. Takayama, C. Wang et al. Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 2005 |
| Interaction | PhysicalInteraction Rv0129c | prabhakarsmail | IPI | | structural analysis
authors,E. Lozes,K. Huygen,J. Content,O. Denis,DL. Montgomery,AM. Yawman,P. Vandenbussche,JP. Van Vooren,A. Drowart,JB. Ulmer,MA. Liu Immunogenicity and efficacy of a tuberculosis DNA vaccine encoding the components of the secreted antigen 85 complex. Vaccine 1997 |
| Interaction | PhysicalInteraction Rv0129c | prabhakarsmail | IPI | | structural analysis
authors,G. Kumar,PK. Dagur,M. Singh,VS. Yadav,R. Dayal,HB. Singh,VM. Katoch,U. Sengupta,B. Joshi Diagnostic potential of Ag85C in comparison to various secretory antigens for childhood tuberculosis. Scand. J. Immunol. 2008 |
| Interaction | PhysicalInteraction Rv0129c | prabhakarsmail | IPI | | structural analysis
authors,N. Matluk,JA. Rochira,A. Karaczyn,T. Adams,JM. Verdi A role for NRAGE in NF-kappaB activation through the non-canonical BMP pathway. BMC Biol. 2010 |
| Interaction | PhysicalInteraction Rv0129c | prabhakarsmail | IPI | | Spectrophometric assay
authors,N. Matluk,JA. Rochira,A. Karaczyn,T. Adams,JM. Verdi A role for NRAGE in NF-kappaB activation through the non-canonical BMP pathway. BMC Biol. 2010 |
| Interaction | PhysicalInteraction Rv0129c | prabhakarsmail | IPI | | Spectrophometric assay
K. Takayama, C. Wang et al. Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 2005 |
| Interaction | PhysicalInteraction Rv0129c | prabhakarsmail | IPI | | structural analysis
authors,DR. Ronning,T. Klabunde,GS. Besra,VD. Vissa,JT. Belisle,JC. Sacchettini Crystal structure of the secreted form of antigen 85C reveals potential targets for mycobacterial drugs and vaccines. Nat. Struct. Biol. 2000 |
| Interaction | PhysicalInteraction Rv0129c | prabhakarsmail | IPI | | Spectrophometric assay
authors,E. Lozes,K. Huygen,J. Content,O. Denis,DL. Montgomery,AM. Yawman,P. Vandenbussche,JP. Van Vooren,A. Drowart,JB. Ulmer,MA. Liu Immunogenicity and efficacy of a tuberculosis DNA vaccine encoding the components of the secreted antigen 85 complex. Vaccine 1997 |
| Interaction | PhysicalInteraction Rv0129c | prabhakarsmail | IPI | | Spectrophometric assay
authors,G. Kumar,PK. Dagur,M. Singh,VS. Yadav,R. Dayal,HB. Singh,VM. Katoch,U. Sengupta,B. Joshi Diagnostic potential of Ag85C in comparison to various secretory antigens for childhood tuberculosis. Scand. J. Immunol. 2008 |
| Interaction | PhysicalInteraction Rv0129c | prabhakarsmail | IPI | | Spectrophometric assay
authors,DR. Ronning,T. Klabunde,GS. Besra,VD. Vissa,JT. Belisle,JC. Sacchettini Crystal structure of the secreted form of antigen 85C reveals potential targets for mycobacterial drugs and vaccines. Nat. Struct. Biol. 2000 |
| Interaction | RegulatedBy Rv0757 | yamir.moreno | IEP | | Microarrays. mRNA levels of regulated element measured and compared between wild-type and trans-element mutation (knockout, over expression etc.) performed by using microarray (or macroarray) experiments.. qRT-PCR. mRNA expression levels of regulated element measured and compared between wild-type and trans-element mutation (knockout, over expression etc.) performed by using qRT-PCR technique.
SB. Walters, E. Dubnau et al. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 2006 |
| Interaction | RegulatedBy Rv0757 | yamir.moreno | IEP | | Microarrays. mRNA levels of regulated element measured and compared between wild-type and trans-element mutation (knockout, over expression etc.) performed by using microarray (or macroarray) experiments.. qRT-PCR. mRNA expression levels of regulated element measured and compared between wild-type and trans-element mutation (knockout, over expression etc.) performed by using qRT-PCR technique.
SB. Walters, E. Dubnau et al. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 2006 |
| Interaction | RegulatedBy Rv1221 | yamir.moreno | IEP | | Microarrays. mRNA levels of regulated element measured and compared between wild-type and trans-element mutation (knockout, over expression etc.) performed by using microarray (or macroarray) experiments..
R. Manganelli, MI. Voskuil et al. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol. Microbiol. 2001 |
| Name | acyl CoA:diacylglycerol acyltransferase | mjackson | IMP | | Triglyceride biosynthesis |
| Name | acyl CoA:diacylglycerol acyltransferase | mjackson | IDA | | Triglyceride biosynthesis |
| Name | mycolyltransferase 85A;INVOLVED IN CELL WALL MYCOLOYLATION | mjackson | IMP | | Mycolic acid processing |
| Name | mycolyltransferase 85A;INVOLVED IN CELL WALL MYCOLOYLATION | mjackson | IDA | | Mycolic acid processing |
| Symbol | Ag85A | jlew | | | Tat substrate. Demonstrated by experimental verification that four secreted proteins from Mtb carrying putative Tat signals are bona fide Tat substrates
M. Marrichi, L. Camacho et al. Genetic Toggling of Alkaline Phosphatase Folding Reveals Signal Peptides for All Major Modes of Transport across the Inner Membrane of Bacteria. J. Biol. Chem. 2008 |
| Citation | Genetic Toggling of Alkaline Phosphatase Folding Reveals Signal Peptides for All Major Modes of Transport across the Inner Membrane of Bacteria. M. Marrichi, L. Camacho et al. J. Biol. Chem. 2008 | jlew | | 18819916 | Tat substrate. Demonstrated by experimental verification that four secreted proteins from Mtb carrying putative Tat signals are bona fide Tat substrates |
| Citation | The Mycobacterium tuberculosis Ag85A is a novel diacylglycerol acyltransferase involved in lipid body formation. authors,AA. Elamin,M. Stehr,R. Spallek,M. Rohde,M. Singh Mol. Microbiol. 2011 | mjackson | | 21819455 | acyl CoA:diacylglycerol acyltransferase (mycobacterial recombinant strain phenotype; enzymatic) |
| Other | TBPWY:Triglyceride biosynthesis | mjackson | | | acyl CoA:diacylglycerol acyltransferase (mycobacterial recombinant strain phenotype; enzymatic)
authors,AA. Elamin,M. Stehr,R. Spallek,M. Rohde,M. Singh The Mycobacterium tuberculosis Ag85A is a novel diacylglycerol acyltransferase involved in lipid body formation. Mol. Microbiol. 2011 |
| Citation | Biosynthesis of the arabinogalactan-peptidoglycan complex of Mycobacterium tuberculosis. authors,DC. Crick,S. Mahapatra,PJ. Brennan Glycobiology 2001 | jjmcfadden | | 11555614 | Inferred from direct assay |
| Term | EC:2.3.1.- Transferases. Acyltransferases. Transferring groups other than amino-acyl groups. - NR | jjmcfadden | NR | | Inferred from direct assay
authors,DC. Crick,S. Mahapatra,PJ. Brennan Biosynthesis of the arabinogalactan-peptidoglycan complex of Mycobacterium tuberculosis. Glycobiology 2001 |
| Term | EC:2.3.1.- Transferases. Acyltransferases. Transferring groups other than amino-acyl groups. - NR | extern:JZUCKER | NR | | Assay of protein purified to homogeneity from its native host
authors,JT. Belisle,VD. Vissa,T. Sievert,K. Takayama,PJ. Brennan,GS. Besra Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 1997 |
| Citation | Evidence for a partial redundancy of the fibronectin-binding proteins for the transfer of mycoloyl residues onto the cell wall arabinogalactan termini of Mycobacterium tuberculosis. V. Puech, C. Guilhot et al. Mol. Microbiol. 2002 | extern:JZUCKER | | 12010501 | Inferred from mutant phenotype |
| Term | EC:2.3.1.122 Trehalose O-mycolyltransferase. - NR | extern:JZUCKER | NR | | Inferred from mutant phenotype
V. Puech, C. Guilhot et al. Evidence for a partial redundancy of the fibronectin-binding proteins for the transfer of mycoloyl residues onto the cell wall arabinogalactan termini of Mycobacterium tuberculosis. Mol. Microbiol. 2002 |
| Term | EC:2.3.1.- Transferases. Acyltransferases. Transferring groups other than amino-acyl groups. - NR | extern:JZUCKER | NR | | Inferred from mutant phenotype
V. Puech, C. Guilhot et al. Evidence for a partial redundancy of the fibronectin-binding proteins for the transfer of mycoloyl residues onto the cell wall arabinogalactan termini of Mycobacterium tuberculosis. Mol. Microbiol. 2002 |
| Citation | Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. authors,JT. Belisle,VD. Vissa,T. Sievert,K. Takayama,PJ. Brennan,GS. Besra Science 1997 | extern:JZUCKER | | 9162010 | Assay of protein purified to homogeneity from its native host |
| Term | EC:2.3.1.122 Trehalose O-mycolyltransferase. - NR | extern:JZUCKER | NR | | Assay of protein purified to homogeneity from its native host
authors,JT. Belisle,VD. Vissa,T. Sievert,K. Takayama,PJ. Brennan,GS. Besra Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 1997 |