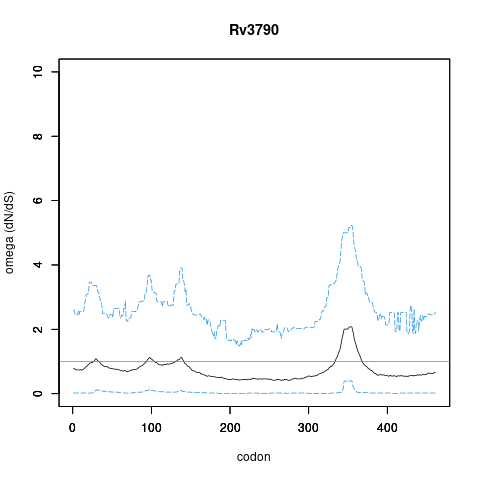
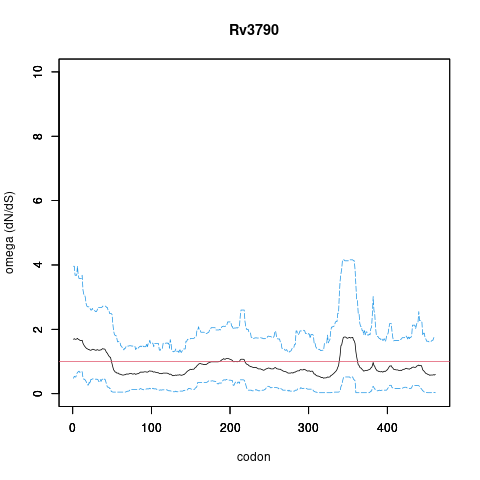
| Property | Value | Creator | Evidence | PMID | Comment |
| Term | TBRXN:DCPE decaprenylphosphoryl ribose, arabinofuranose isomerization - IDA | njamshidi | IDA | 16291675 | see PMID: 16291675. lumped reaction, believed to be two step epimerization of ribose to arabinofuranose
K. Mikusov, H. Huang et al. Decaprenylphosphoryl arabinofuranose, the donor of the D-arabinofuranosyl residues of mycobacterial arabinan, is formed via a two-step epimerization of decaprenylphosphoryl ribose. J. Bacteriol. 2005 |
| Citation | Decaprenylphosphoryl arabinofuranose, the donor of the D-arabinofuranosyl residues of mycobacterial arabinan, is formed via a two-step epimerization of decaprenylphosphoryl ribose. K. Mikusov, H. Huang et al. J. Bacteriol. 2005 | njamshidi | IPI | 16291675 | see PMID: 16291675. lumped reaction, believed to be two step epimerization of ribose to arabinofuranose |
| Term | TBRXN:DCPE decaprenylphosphoryl ribose, arabinofuranose isomerization - IPI | njamshidi | IPI | 16291675 | see PMID: 16291675. lumped reaction, believed to be two step epimerization of ribose to arabinofuranose
K. Mikusov, H. Huang et al. Decaprenylphosphoryl arabinofuranose, the donor of the D-arabinofuranosyl residues of mycobacterial arabinan, is formed via a two-step epimerization of decaprenylphosphoryl ribose. J. Bacteriol. 2005 |
| Citation | Decaprenylphosphoryl arabinofuranose, the donor of the D-arabinofuranosyl residues of mycobacterial arabinan, is formed via a two-step epimerization of decaprenylphosphoryl ribose. K. Mikusov, H. Huang et al. J. Bacteriol. 2005 | njamshidi | IDA | 16291675 | see PMID: 16291675. lumped reaction, believed to be two step epimerization of ribose to arabinofuranose |
| Symbol | dprE1 | mjackson | IDA | | Decaprenyl phosphoarabinose synthesis |
| Name | decaprenylphosphoryl-2-keto-beta-D-erythro-pentofuranose synthase (AKA Decaprenylphosphoryl-D-ribose oxidase), the first step in epimerizing decaprenyl ribose to decaprenyl arabinose | mjackson | IDA | | Decaprenyl phosphoarabinose synthesis |
| Symbol | dprE1 | mmcneil | | | decaprenylphosphoryl-2-keto-beta-D-erythro-pentofuranose synthase (AKA Decaprenylphosphoryl-D-ribose oxidase), the first step in epimerizing decaprenyl ribose to decaprenyl arabinose; enzymatic evidence
K. Mikusov, H. Huang et al. Decaprenylphosphoryl arabinofuranose, the donor of the D-arabinofuranosyl residues of mycobacterial arabinan, is formed via a two-step epimerization of decaprenylphosphoryl ribose. J. Bacteriol. 2005 |
| Citation | Decaprenylphosphoryl arabinofuranose, the donor of the D-arabinofuranosyl residues of mycobacterial arabinan, is formed via a two-step epimerization of decaprenylphosphoryl ribose. K. Mikusov, H. Huang et al. J. Bacteriol. 2005 | mmcneil | | 16291675 | decaprenylphosphoryl-2-keto-beta-D-erythro-pentofuranose synthase (AKA Decaprenylphosphoryl-D-ribose oxidase), the first step in epimerizing decaprenyl ribose to decaprenyl arabinose; enzymatic evidence |
| Term | GO:0009247 glycolipid biosynthetic process - NR | mmcneil | NR | | decaprenylphosphoryl-2-keto-beta-D-erythro-pentofuranose synthase (AKA Decaprenylphosphoryl-D-ribose oxidase), the first step in epimerizing decaprenyl ribose to decaprenyl arabinose; enzymatic evidence
K. Mikusov, H. Huang et al. Decaprenylphosphoryl arabinofuranose, the donor of the D-arabinofuranosyl residues of mycobacterial arabinan, is formed via a two-step epimerization of decaprenylphosphoryl ribose. J. Bacteriol. 2005 |
| Citation | Identification and active expression of the Mycobacterium tuberculosis gene encoding 5-phospho-{alpha}-d-ribose-1-diphosphate: decaprenyl-phosphate 5-phosphoribosyltransferase, the first enzyme committed to decaprenylphosphoryl-d-arabinose synthesis. H. Huang, MS. Scherman et al. J. Biol. Chem. 2005 | jjmcfadden | | 15878857 | Inferred from direct assay |
| Term | EC:1.-.-.- Oxidoreductases. - NR | jjmcfadden | NR | | Inferred from direct assay
H. Huang, MS. Scherman et al. Identification and active expression of the Mycobacterium tuberculosis gene encoding 5-phospho-{alpha}-d-ribose-1-diphosphate: decaprenyl-phosphate 5-phosphoribosyltransferase, the first enzyme committed to decaprenylphosphoryl-d-arabinose synthesis. J. Biol. Chem. 2005 |
| Citation | Identification and active expression of the Mycobacterium tuberculosis gene encoding 5-phospho-{alpha}-d-ribose-1-diphosphate: decaprenyl-phosphate 5-phosphoribosyltransferase, the first enzyme committed to decaprenylphosphoryl-d-arabinose synthesis. H. Huang, MS. Scherman et al. J. Biol. Chem. 2005 | extern:JZUCKER | | 15878857 | Inferred from direct assay |
| Term | EC:1.-.-.- Oxidoreductases. - NR | extern:JZUCKER | NR | | Inferred from direct assay
H. Huang, MS. Scherman et al. Identification and active expression of the Mycobacterium tuberculosis gene encoding 5-phospho-{alpha}-d-ribose-1-diphosphate: decaprenyl-phosphate 5-phosphoribosyltransferase, the first enzyme committed to decaprenylphosphoryl-d-arabinose synthesis. J. Biol. Chem. 2005 |