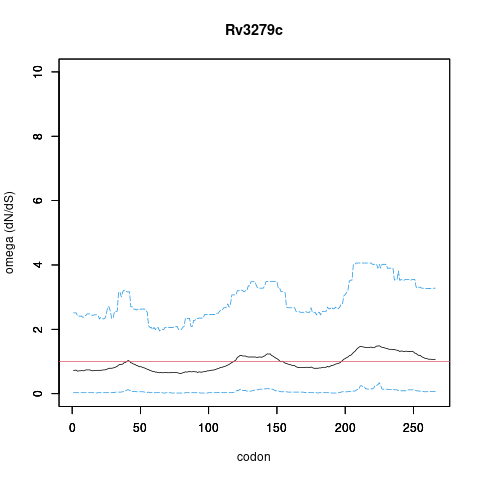
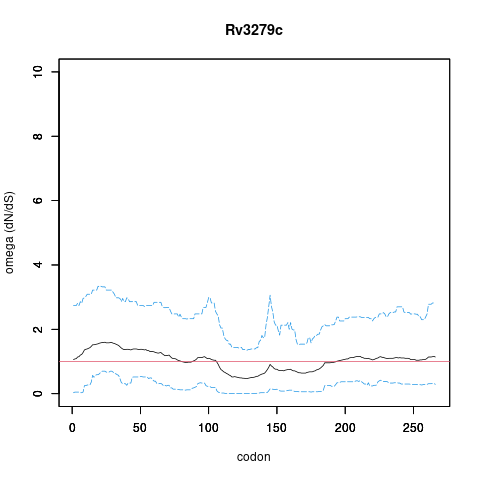
| Property | Value | Creator | Evidence | PMID | Comment |
| Interaction | Regulates Rv1568 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Citation | The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. G. Balzsi, AP. Heath et al. Mol. Syst. Biol. 2008 | yamir.moreno | ISO | 18985025 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Interaction | Regulates Rv1589 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Citation | The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. G. Balzsi, AP. Heath et al. Mol. Syst. Biol. 2008 | yamir.moreno | ISO | 18985025 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Interaction | Regulates Rv0089 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Citation | The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. G. Balzsi, AP. Heath et al. Mol. Syst. Biol. 2008 | yamir.moreno | ISO | 18985025 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Interaction | Regulates Rv1176c | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Citation | The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. G. Balzsi, AP. Heath et al. Mol. Syst. Biol. 2008 | yamir.moreno | ISO | 18985025 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Citation | Evolutionary dynamics of prokaryotic transcriptional regulatory networks. authors,M. Madan Babu,SA. Teichmann,L. Aravind J. Mol. Biol. 2006 | yamir.moreno | ISO | 16530225 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Interaction | Regulates Rv1176c | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
authors,M. Madan Babu,SA. Teichmann,L. Aravind Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006 |
| Citation | Evolutionary dynamics of prokaryotic transcriptional regulatory networks. authors,M. Madan Babu,SA. Teichmann,L. Aravind J. Mol. Biol. 2006 | yamir.moreno | ISO | 16530225 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Interaction | Regulates Rv1568 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
authors,M. Madan Babu,SA. Teichmann,L. Aravind Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006 |
| Citation | Evolutionary dynamics of prokaryotic transcriptional regulatory networks. authors,M. Madan Babu,SA. Teichmann,L. Aravind J. Mol. Biol. 2006 | yamir.moreno | ISO | 16530225 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Interaction | Regulates Rv1589 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
authors,M. Madan Babu,SA. Teichmann,L. Aravind Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006 |
| Citation | Evolutionary dynamics of prokaryotic transcriptional regulatory networks. authors,M. Madan Babu,SA. Teichmann,L. Aravind J. Mol. Biol. 2006 | yamir.moreno | ISO | 16530225 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Interaction | Regulates Rv0089 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
authors,M. Madan Babu,SA. Teichmann,L. Aravind Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006 |