| Property | Value | Creator | Evidence | PMID | Comment |
| Citation | Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. authors,J. Daniel,H. Maamar,C. Deb,TD. Sirakova,PE. Kolattukudy PLoS Pathog. 2011 | jlew | | 21731490 | Among genes upregulated in Mtb in hypoxic lipid loaded macrophages. |
| Symbol | lipY | jlew | | | The role of the PE domain in ESX-5 secretion was confirmed in a whole cell lipase assay, in which wild-type bacteria expressing full-length LipYtub, but not LipYtub lacking its PE domain, were shown to hydrolyze extracellular lipids. In conclusion, both PE and PPE domains contain a signal required for secretion of LipY by the ESX-5 system, and these domains are proteolytically removed upon translocation.
authors,MH. Daleke,A. Cascioferro,K. de Punder,R. Ummels,AM. Abdallah,N. van der Wel,PJ. Peters,J. Luirink,R. Manganelli,W. Bitter Conserved Pro-Glu (PE) and Pro-Pro-Glu (PPE) protein domains target LipY lipases of pathogenic mycobacteria to the cell surface via the ESX-5 pathway. J. Biol. Chem. 2011 |
| Citation | Conserved Pro-Glu (PE) and Pro-Pro-Glu (PPE) protein domains target LipY lipases of pathogenic mycobacteria to the cell surface via the ESX-5 pathway. authors,MH. Daleke,A. Cascioferro,K. de Punder,R. Ummels,AM. Abdallah,N. van der Wel,PJ. Peters,J. Luirink,R. Manganelli,W. Bitter J. Biol. Chem. 2011 | jlew | | 21471225 | The role of the PE domain in ESX-5 secretion was confirmed in a whole cell lipase assay, in which wild-type bacteria expressing full-length LipYtub, but not LipYtub lacking its PE domain, were shown to hydrolyze extracellular lipids. In conclusion, both PE and PPE domains contain a signal required for secretion of LipY by the ESX-5 system, and these domains are proteolytically removed upon translocation. |
| Symbol | lipY | jlew | | | Rv3097c of Mycobacterium tuberculosis encoding lipase (LipY) was overexpressed in Mycobacterium bovis BCG. Immunogenicity of BCG vaccine was impaired by LipY-induced hydrolysis of specific lipids leading to suppression of host immune responses.
authors,VK. Singh,V. Srivastava,V. Singh,N. Rastogi,R. Roy,AK. Shaw,AK. Dwivedi,R. Srivastava,BS. Srivastava Overexpression of Rv3097c in Mycobacterium bovis BCG abolished the efficacy of BCG vaccine to protect against Mycobacterium tuberculosis infection in mice. Vaccine 2011 |
| Citation | Overexpression of Rv3097c in Mycobacterium bovis BCG abolished the efficacy of BCG vaccine to protect against Mycobacterium tuberculosis infection in mice. authors,VK. Singh,V. Srivastava,V. Singh,N. Rastogi,R. Roy,AK. Shaw,AK. Dwivedi,R. Srivastava,BS. Srivastava Vaccine 2011 | jlew | | 21565242 | Rv3097c of Mycobacterium tuberculosis encoding lipase (LipY) was overexpressed in Mycobacterium bovis BCG. Immunogenicity of BCG vaccine was impaired by LipY-induced hydrolysis of specific lipids leading to suppression of host immune responses. |
| Symbol | lipY | jlew | | | Among genes upregulated in Mtb in hypoxic lipid loaded macrophages.
authors,J. Daniel,H. Maamar,C. Deb,TD. Sirakova,PE. Kolattukudy Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog. 2011 |
| Symbol | PE_PGRS/LipY | jlew | | | interacts with Rv2763c and Rv3733c. Using an in vivo pull-down assay, for the first time we have confirmed the presence of three pairs of PE/PPE-related novel protein interactions in this pathogen.
J. Zeng, L. Zhang et al. Over-producing soluble protein complex and validating protein-protein interaction through a new bacterial co-expression system. Protein Expr. Purif. 2010 |
| Citation | Over-producing soluble protein complex and validating protein-protein interaction through a new bacterial co-expression system. J. Zeng, L. Zhang et al. Protein Expr. Purif. 2010 | jlew | | 19747546 | interacts with Rv2763c and Rv3733c. Using an in vivo pull-down assay, for the first time we have confirmed the presence of three pairs of PE/PPE-related novel protein interactions in this pathogen. |
| Symbol | lipY | jlew | | | We have characterized the triacylglycerol (TAG) hydrolase LipY from Mycobacterium tuberculosis. Results suggest that the PE domain modulates enzyme activity.
authors,KC. Mishra,C. de Chastellier,Y. Narayana,P. Bifani,AK. Brown,GS. Besra,VM. Katoch,B. Joshi,KN. Balaji,L. Kremer Functional role of the PE domain and immunogenicity of the Mycobacterium tuberculosis triacylglycerol hydrolase LipY. Infect. Immun. 2008 |
| Citation | Functional role of the PE domain and immunogenicity of the Mycobacterium tuberculosis triacylglycerol hydrolase LipY. authors,KC. Mishra,C. de Chastellier,Y. Narayana,P. Bifani,AK. Brown,GS. Besra,VM. Katoch,B. Joshi,KN. Balaji,L. Kremer Infect. Immun. 2008 | jlew | | 17938218 | We have characterized the triacylglycerol (TAG) hydrolase LipY from Mycobacterium tuberculosis. Results suggest that the PE domain modulates enzyme activity. |
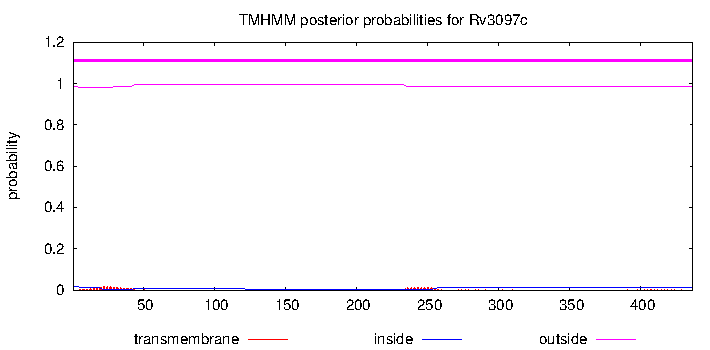
| Citation | PE is a functional domain responsible for protein translocation and localization on mycobacterial cell wall. A. Cascioferro, G. Delogu et al. Mol. Microbiol. 2007 | jlew | | 18028308 | Protein is surface exposed and localizes in the cell wall. |
| Citation | Macrophage-specific Mycobacterium tuberculosis genes: identification by green fluorescent protein and kanamycin resistance selection. V. Srivastava, C. Rouanet et al. Microbiology (Reading, Engl.) 2007 | jlew | | 17322185 | Reporter finds TB promoters that are upregulated in macrophages compared to in vitro growth. |
| Citation | A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. C. Deb, J. Daniel et al. J. Biol. Chem. 2006 | jlew | | 16354661 | We show that Rv3097c (LIPY) hydrolyzed long-chain TG with high specific activity. We demonstrate that hypoxic cultures of M. tuberculosis, which had accumulated TG, hydrolyzed the stored TG when subjected to nutrient starvation. Under such conditions, lipY was induced more than all lipases, suggesting a central role for it in the utilization of stored TG. We also show that in the lipY-deficient mutant, TG utilization was drastically decreased under nutrient deprived condition. |
| Symbol | lipY | jlew | | | We show that Rv3097c (LIPY) hydrolyzed long-chain TG with high specific activity. We demonstrate that hypoxic cultures of M. tuberculosis, which had accumulated TG, hydrolyzed the stored TG when subjected to nutrient starvation. Under such conditions, lipY was induced more than all lipases, suggesting a central role for it in the utilization of stored TG. We also show that in the lipY-deficient mutant, TG utilization was drastically decreased under nutrient deprived condition.
C. Deb, J. Daniel et al. A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J. Biol. Chem. 2006 |
| Citation | A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. C. Deb, J. Daniel et al. J. Biol. Chem. 2006 | extern:JZUCKER | | 16354661 | Assay of protein purified to homogeneity from its native host |
| Term | EC:3.1.1.3 Triacylglycerol lipase. - NR | extern:JZUCKER | NR | | Assay of protein purified to homogeneity from its native host
C. Deb, J. Daniel et al. A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J. Biol. Chem. 2006 |