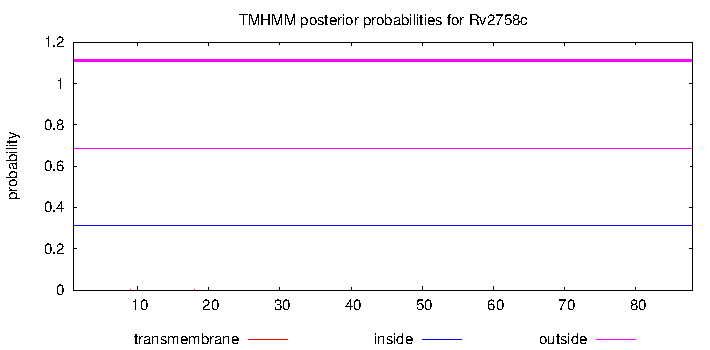
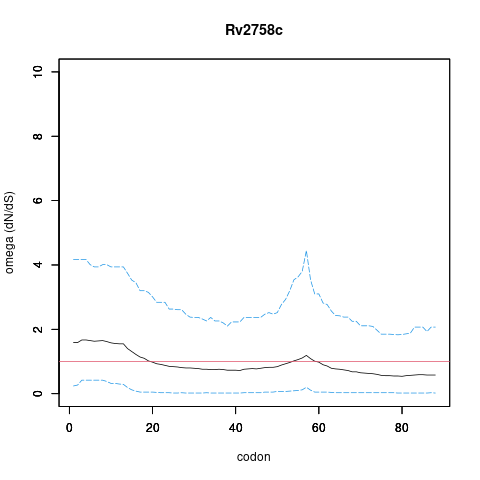
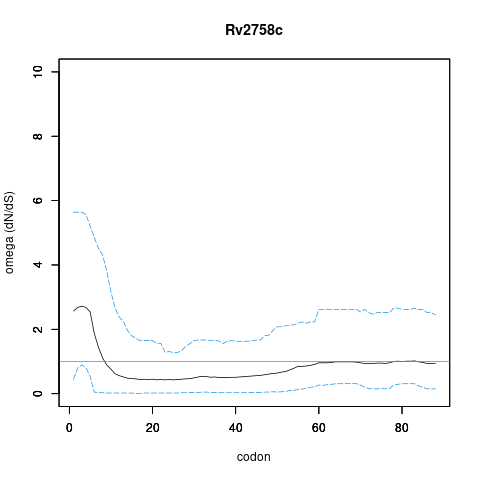
| Property | Value | Creator | Evidence | PMID | Comment |
| Interaction | Inhibits Rv2757c | vizhi.gurusamy | IEP | | Co-expression (Functional linkage)
authors,VL. Arcus,PB. Rainey,SJ. Turner The PIN-domain toxin-antitoxin array in mycobacteria. Trends Microbiol. 2005 |
| Interaction | Inhibits Rv2757c | vizhi.gurusamy | IEP | | Co-expression (Functional linkage)
authors,A. Gupta Killing activity and rescue function of genome-wide toxin-antitoxin loci of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 2009 |
| Citation | Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. authors,DP. Pandey,K. Gerdes Nucleic Acids Res. 2005 | vizhi.gurusamy | IEP | 15718296 | Co-expression (Functional linkage) |
| Interaction | Inhibits Rv2757c | vizhi.gurusamy | IEP | | Co-expression (Functional linkage)
authors,DP. Pandey,K. Gerdes Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005 |
| Citation | The PIN-domain toxin-antitoxin array in mycobacteria. authors,VL. Arcus,PB. Rainey,SJ. Turner Trends Microbiol. 2005 | vizhi.gurusamy | IEP | 15993073 | Co-expression (Functional linkage) |
| Interaction | Inhibits Rv2757c | vizhi.gurusamy | IEP | | Co-expression (Functional linkage)
authors,VL. Arcus,PB. Rainey,SJ. Turner The PIN-domain toxin-antitoxin array in mycobacteria. Trends Microbiol. 2005 |
| Symbol | VapB21 | jlew | | | We report the heterologous toxicity of these TA loci in Escherichia coli and show that only a few of the M. tuberculosis-encoded toxins can inhibit E. coli growth and have a killing effect. This killing effect can be suppressed by coexpression of the cognate antitoxin.
authors,A. Gupta Killing activity and rescue function of genome-wide toxin-antitoxin loci of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 2009 |
| Citation | Killing activity and rescue function of genome-wide toxin-antitoxin loci of Mycobacterium tuberculosis. authors,A. Gupta FEMS Microbiol. Lett. 2009 | jlew | | 19016878 | We report the heterologous toxicity of these TA loci in Escherichia coli and show that only a few of the M. tuberculosis-encoded toxins can inhibit E. coli growth and have a killing effect. This killing effect can be suppressed by coexpression of the cognate antitoxin. |
| Other | stop:3070849 | rslayden | | | MTCY48.05c (72 aa), FASTA scores: opt: 106, E(): 0.52, (39.15% identity in 46 aa overlap); O06565 |
| Other | strand:+ | rslayden | | | MTCY48.05c (72 aa), FASTA scores: opt: 106, E(): 0.52, (39.15% identity in 46 aa overlap); O06565 |
| Symbol | Vap BC-21 | rslayden | | | Rv1113 |
| Other | start:3070583 | rslayden | | | Rv1113 |
| Other | stop:3070849 | rslayden | | | Rv1113 |
| Other | strand:+ | rslayden | | | Rv1113 |
| Symbol | Vap BC-21 | rslayden | | | MTCY22G8.02 (65 aa), FASTA scores: opt: 97, E(): 2.2, (33.35% identity in 69 aa overlap); etc. Contains PS00402 Binding-protein-dependent transport systems inner membrane comp signature. |
| Other | start:3070583 | rslayden | | | MTCY22G8.02 (65 aa), FASTA scores: opt: 97, E(): 2.2, (33.35% identity in 69 aa overlap); etc. Contains PS00402 Binding-protein-dependent transport systems inner membrane comp signature. |
| Other | stop:3070849 | rslayden | | | MTCY22G8.02 (65 aa), FASTA scores: opt: 97, E(): 2.2, (33.35% identity in 69 aa overlap); etc. Contains PS00402 Binding-protein-dependent transport systems inner membrane comp signature. |
| Other | strand:+ | rslayden | | | MTCY22G8.02 (65 aa), FASTA scores: opt: 97, E(): 2.2, (33.35% identity in 69 aa overlap); etc. Contains PS00402 Binding-protein-dependent transport systems inner membrane comp signature. |
| Symbol | Vap BC-21 | rslayden | | | Conserved hypothetical protein, similar to several other M. tuberculosis hypothetical proteins e.g. P95008 |
| Other | start:3070583 | rslayden | | | Conserved hypothetical protein, similar to several other M. tuberculosis hypothetical proteins e.g. P95008 |
| Other | stop:3070849 | rslayden | | | Conserved hypothetical protein, similar to several other M. tuberculosis hypothetical proteins e.g. P95008 |
| Other | strand:+ | rslayden | | | Conserved hypothetical protein, similar to several other M. tuberculosis hypothetical proteins e.g. P95008 |
| Symbol | Vap BC-21 | rslayden | | | Rv2545 (92 aa), FASTA scores: opt: 151, E(): 0.00028, (66.65%identity in 45 aa overlap); Q10771 |
| Other | start:3070583 | rslayden | | | Rv2545 (92 aa), FASTA scores: opt: 151, E(): 0.00028, (66.65%identity in 45 aa overlap); Q10771 |
| Other | stop:3070849 | rslayden | | | Rv2545 (92 aa), FASTA scores: opt: 151, E(): 0.00028, (66.65%identity in 45 aa overlap); Q10771 |
| Other | strand:+ | rslayden | | | Rv2545 (92 aa), FASTA scores: opt: 151, E(): 0.00028, (66.65%identity in 45 aa overlap); Q10771 |
| Symbol | Vap BC-21 | rslayden | | | YF60_MYCTU |
| Other | start:3070583 | rslayden | | | YF60_MYCTU |
| Other | stop:3070849 | rslayden | | | YF60_MYCTU |
| Other | strand:+ | rslayden | | | YF60_MYCTU |
| Symbol | Vap BC-21 | rslayden | | | Rv1560 |
| Other | start:3070583 | rslayden | | | Rv1560 |
| Other | stop:3070849 | rslayden | | | Rv1560 |
| Other | strand:+ | rslayden | | | Rv1560 |
| Symbol | Vap BC-21 | rslayden | | | MT1611 |
| Other | start:3070583 | rslayden | | | MT1611 |
| Other | stop:3070849 | rslayden | | | MT1611 |
| Other | strand:+ | rslayden | | | MT1611 |
| Symbol | Vap BC-21 | rslayden | | | MTCY48.05c (72 aa), FASTA scores: opt: 106, E(): 0.52, (39.15% identity in 46 aa overlap); O06565 |
| Other | start:3070583 | rslayden | | | MTCY48.05c (72 aa), FASTA scores: opt: 106, E(): 0.52, (39.15% identity in 46 aa overlap); O06565 |