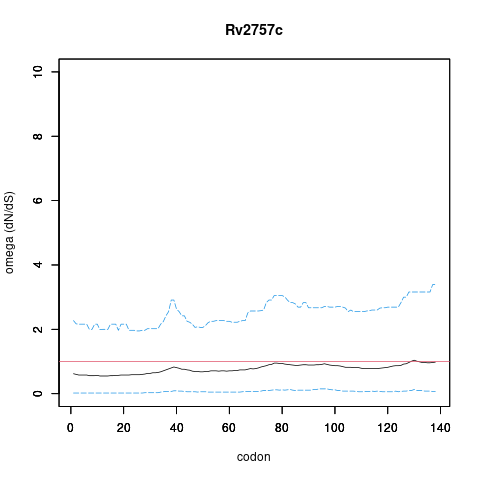
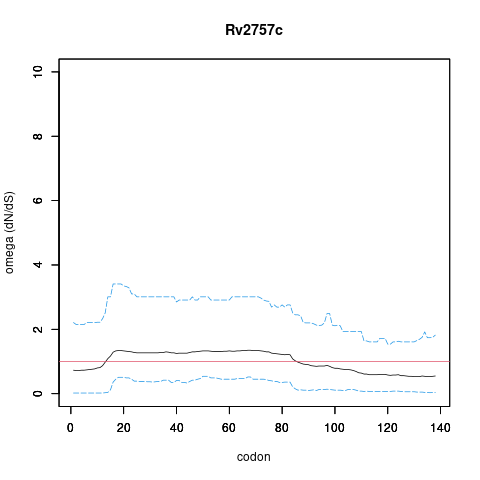
| Property | Value | Creator | Evidence | PMID | Comment |
| Citation | The PIN-domain toxin-antitoxin array in mycobacteria. authors,VL. Arcus,PB. Rainey,SJ. Turner Trends Microbiol. 2005 | vizhi.gurusamy | IEP | 15993073 | Co-expression (Functional linkage) |
| Interaction | InhibitedBy Rv2758c | vizhi.gurusamy | IEP | | Co-expression (Functional linkage)
authors,VL. Arcus,PB. Rainey,SJ. Turner The PIN-domain toxin-antitoxin array in mycobacteria. Trends Microbiol. 2005 |
| Citation | Killing activity and rescue function of genome-wide toxin-antitoxin loci of Mycobacterium tuberculosis. authors,A. Gupta FEMS Microbiol. Lett. 2009 | vizhi.gurusamy | IEP | 19016878 | Co-expression (Functional linkage) |
| Interaction | InhibitedBy Rv2758c | vizhi.gurusamy | IEP | | Co-expression (Functional linkage)
authors,A. Gupta Killing activity and rescue function of genome-wide toxin-antitoxin loci of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 2009 |
| Interaction | InhibitedBy Rv2758c | vizhi.gurusamy | IEP | | Co-expression (Functional linkage)
authors,DP. Pandey,K. Gerdes Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005 |
| Interaction | InhibitedBy Rv2758c | vizhi.gurusamy | IEP | | Co-expression (Functional linkage)
authors,VL. Arcus,PB. Rainey,SJ. Turner The PIN-domain toxin-antitoxin array in mycobacteria. Trends Microbiol. 2005 |
| Interaction | Regulatory Rv0616c | akankshajain.21 | IEP | | Co-expression (Functional linkage)
T. Parish, DA. Smith et al. The senX3-regX3 two-component regulatory system of Mycobacterium tuberculosis is required for virulence. Microbiology (Reading, Engl.) 2003 |
| Interaction | RegulatedBy Rv0491 | yamir.moreno | IEP | | Microarrays. mRNA levels of regulated element measured and compared between wild-type and trans-element mutation (knockout, over expression etc.) performed by using microarray (or macroarray) experiments..
T. Parish, DA. Smith et al. The senX3-regX3 two-component regulatory system of Mycobacterium tuberculosis is required for virulence. Microbiology (Reading, Engl.) 2003 |
| Citation | Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. authors,HR. Ramage,LE. Connolly,JS. Cox PLoS Genet. 2009 | jlew | | 20011113 | VapC homolog, PIN domain, Toxic when expressed in Msmeg, rescued by antitoxin (Rv2758c) |
| Symbol | VapC21 | jlew | | | We report the heterologous toxicity of these TA loci in Escherichia coli and show that only a few of the M. tuberculosis-encoded toxins can inhibit E. coli growth and have a killing effect. This killing effect can be suppressed by coexpression of the cognate antitoxin.
authors,A. Gupta Killing activity and rescue function of genome-wide toxin-antitoxin loci of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 2009 |
| Citation | Killing activity and rescue function of genome-wide toxin-antitoxin loci of Mycobacterium tuberculosis. authors,A. Gupta FEMS Microbiol. Lett. 2009 | jlew | | 19016878 | We report the heterologous toxicity of these TA loci in Escherichia coli and show that only a few of the M. tuberculosis-encoded toxins can inhibit E. coli growth and have a killing effect. This killing effect can be suppressed by coexpression of the cognate antitoxin. |
| Other | stop:3070586 | rslayden | | | MTCY159.10c (137 aa), FASTA scores: opt: 265, E(): 7.5e-12, (38.5% identity in 135 aa overlap); O07228 |
| Other | strand:+ | rslayden | | | MTCY159.10c (137 aa), FASTA scores: opt: 265, E(): 7.5e-12, (38.5% identity in 135 aa overlap); O07228 |
| Other | start:3070170 | rslayden | | | Rv0301 |
| Other | stop:3070586 | rslayden | | | Rv0301 |
| Other | strand:+ | rslayden | | | Rv0301 |
| Other | start:3070170 | rslayden | | | MTCY63.06 (141 aa), FASTA scores: opt: 259, E(): 2.1e-11, (42.4% identity in 132 aa overlap); etc. |
| Other | stop:3070586 | rslayden | | | MTCY63.06 (141 aa), FASTA scores: opt: 259, E(): 2.1e-11, (42.4% identity in 132 aa overlap); etc. |
| Other | strand:+ | rslayden | | | MTCY63.06 (141 aa), FASTA scores: opt: 259, E(): 2.1e-11, (42.4% identity in 132 aa overlap); etc. |
| Other | start:3070170 | rslayden | | | Conserved hypothetical protein, similar to several other M. tuberculosis hypothetical proteins e.g. P96411 |
| Other | stop:3070586 | rslayden | | | Conserved hypothetical protein, similar to several other M. tuberculosis hypothetical proteins e.g. P96411 |
| Other | strand:+ | rslayden | | | Conserved hypothetical protein, similar to several other M. tuberculosis hypothetical proteins e.g. P96411 |
| Other | start:3070170 | rslayden | | | Rv0229c |
| Other | stop:3070586 | rslayden | | | Rv0229c |
| Other | strand:+ | rslayden | | | Rv0229c |
| Other | start:3070170 | rslayden | | | MTCY08D5.24c (226 aa), FASTA scores: opt: 354, E():4.6e-18, (45.25% identity in 137 aa overlap) (N-terminus longer 89 aa); P95007 |
| Other | stop:3070586 | rslayden | | | MTCY08D5.24c (226 aa), FASTA scores: opt: 354, E():4.6e-18, (45.25% identity in 137 aa overlap) (N-terminus longer 89 aa); P95007 |
| Other | strand:+ | rslayden | | | MTCY08D5.24c (226 aa), FASTA scores: opt: 354, E():4.6e-18, (45.25% identity in 137 aa overlap) (N-terminus longer 89 aa); P95007 |
| Other | start:3070170 | rslayden | | | Rv2546 |
| Other | stop:3070586 | rslayden | | | Rv2546 |
| Other | strand:+ | rslayden | | | Rv2546 |
| Other | start:3070170 | rslayden | | | MTCY159.10c (137 aa), FASTA scores: opt: 265, E(): 7.5e-12, (38.5% identity in 135 aa overlap); O07228 |