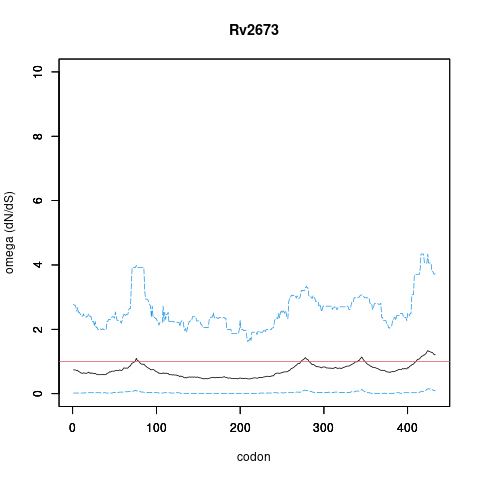
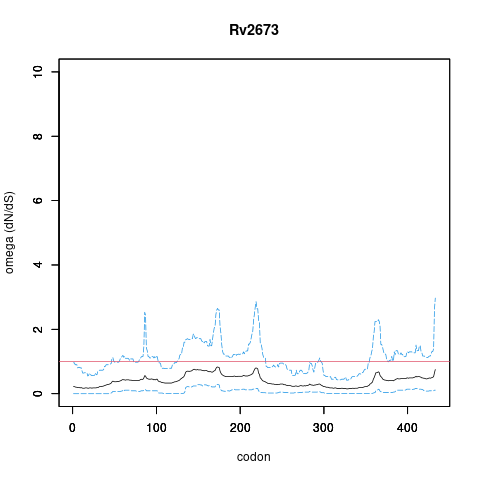
| Property | Value | Creator | Evidence | PMID | Comment |
| Symbol | aftC | mjackson | IMP | | PIM, LM and LAM biosynthesis |
| Name | Decaprenol phosphoarabinose-dependent alpha-1,3-branching arabinosyltransferase involved in the biosynthesis of arabinogalactan and LAM | mjackson | IMP | | PIM, LM and LAM biosynthesis |
| Symbol | aftC | mjackson | IDA | | PIM, LM and LAM biosynthesis |
| Name | Decaprenol phosphoarabinose-dependent alpha-1,3-branching arabinosyltransferase involved in the biosynthesis of arabinogalactan and LAM | mjackson | IDA | | PIM, LM and LAM biosynthesis |
| Symbol | aftC | mjackson | | | Decaprenol phosphoarabinose-dependent alpha-1,3-branching arabinosyltransferase involved in the biosynthesis of arabinogalactan and LAM (phenotypic [mycobacterial recombinant strains]; enzymatic)
HL. Birch,LJ. Alderwick,BJ. Appelmelk,J. Maaskant,A. Bhatt,A. Singh,J. Nigou,L. Eggeling,J. Geurtsen,GS. Besra A truncated lipoglycan from mycobacteria with altered immunological properties. Proc. Natl. Acad. Sci. U.S.A. 2010 |
| Citation | A truncated lipoglycan from mycobacteria with altered immunological properties. HL. Birch,LJ. Alderwick,BJ. Appelmelk,J. Maaskant,A. Bhatt,A. Singh,J. Nigou,L. Eggeling,J. Geurtsen,GS. Besra Proc. Natl. Acad. Sci. U.S.A. 2010 | mjackson | | 20133807 | Decaprenol phosphoarabinose-dependent alpha-1,3-branching arabinosyltransferase involved in the biosynthesis of arabinogalactan and LAM (phenotypic [mycobacterial recombinant strains]; enzymatic) |
| Other | TBPWY:PIM, LM and LAM biosynthesis | mjackson | | | Decaprenol phosphoarabinose-dependent alpha-1,3-branching arabinosyltransferase involved in the biosynthesis of arabinogalactan and LAM (phenotypic [mycobacterial recombinant strains]; enzymatic)
HL. Birch,LJ. Alderwick,BJ. Appelmelk,J. Maaskant,A. Bhatt,A. Singh,J. Nigou,L. Eggeling,J. Geurtsen,GS. Besra A truncated lipoglycan from mycobacteria with altered immunological properties. Proc. Natl. Acad. Sci. U.S.A. 2010 |
| Symbol | aftC | mjackson | | | Decaprenol phosphoarabinose-dependent alpha-1,3-branching arabinosyltransferase involved in the biosynthesis of arabinogalactan and LAM (phenotypic [mycobacterial recombinant strains]; enzymatic)
HL. Birch, LJ. Alderwick et al. Biosynthesis of mycobacterial arabinogalactan: identification of a novel alpha(1-->3) arabinofuranosyltransferase. Mol. Microbiol. 2008 |
| Citation | Biosynthesis of mycobacterial arabinogalactan: identification of a novel alpha(1-->3) arabinofuranosyltransferase. HL. Birch, LJ. Alderwick et al. Mol. Microbiol. 2008 | mjackson | | 18627460 | Decaprenol phosphoarabinose-dependent alpha-1,3-branching arabinosyltransferase involved in the biosynthesis of arabinogalactan and LAM (phenotypic [mycobacterial recombinant strains]; enzymatic) |
| Other | TBPWY:PIM, LM and LAM biosynthesis | mjackson | | | Decaprenol phosphoarabinose-dependent alpha-1,3-branching arabinosyltransferase involved in the biosynthesis of arabinogalactan and LAM (phenotypic [mycobacterial recombinant strains]; enzymatic)
HL. Birch, LJ. Alderwick et al. Biosynthesis of mycobacterial arabinogalactan: identification of a novel alpha(1-->3) arabinofuranosyltransferase. Mol. Microbiol. 2008 |
| Symbol | aftC | mjackson | | | Decaprenol phosphoarabinose-dependent alpha-1,3-branching arabinosyltransferase involved in the biosynthesis of arabinogalactan and LAM (phenotypic [mycobacterial recombinant strains]; enzymatic)
authors,J. Zhang,SK. Angala,PK. Pramanik,K. Li,DC. Crick,A. Liav,A. Jozwiak,E. Swiezewska,M. Jackson,D. Chatterjee Reconstitution of functional mycobacterial arabinosyltransferase AftC proteoliposome and assessment of decaprenylphosphorylarabinose analogues as arabinofuranosyl donors. ACS Chem. Biol. 2011 |
| Citation | Reconstitution of functional mycobacterial arabinosyltransferase AftC proteoliposome and assessment of decaprenylphosphorylarabinose analogues as arabinofuranosyl donors. authors,J. Zhang,SK. Angala,PK. Pramanik,K. Li,DC. Crick,A. Liav,A. Jozwiak,E. Swiezewska,M. Jackson,D. Chatterjee ACS Chem. Biol. 2011 | mjackson | | 21595486 | Decaprenol phosphoarabinose-dependent alpha-1,3-branching arabinosyltransferase involved in the biosynthesis of arabinogalactan and LAM (phenotypic [mycobacterial recombinant strains]; enzymatic) |
| Other | TBPWY:PIM, LM and LAM biosynthesis | mjackson | | | Decaprenol phosphoarabinose-dependent alpha-1,3-branching arabinosyltransferase involved in the biosynthesis of arabinogalactan and LAM (phenotypic [mycobacterial recombinant strains]; enzymatic)
authors,J. Zhang,SK. Angala,PK. Pramanik,K. Li,DC. Crick,A. Liav,A. Jozwiak,E. Swiezewska,M. Jackson,D. Chatterjee Reconstitution of functional mycobacterial arabinosyltransferase AftC proteoliposome and assessment of decaprenylphosphorylarabinose analogues as arabinofuranosyl donors. ACS Chem. Biol. 2011 |