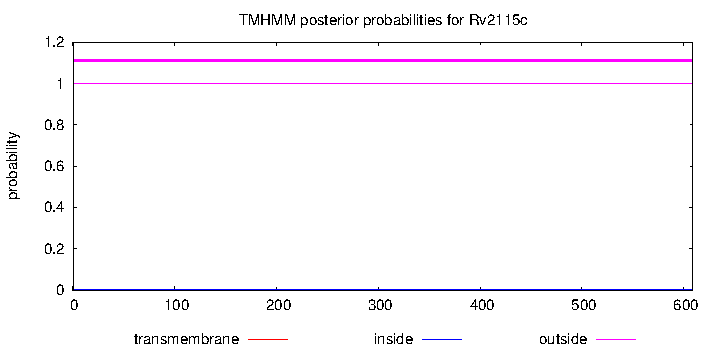
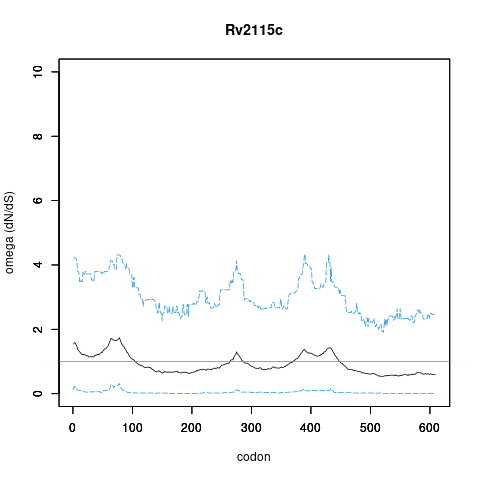
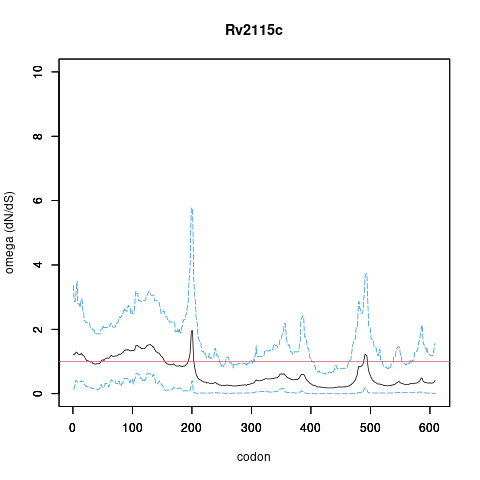
| Property | Value | Creator | Evidence | PMID | Comment |
| Citation | The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. KH. Darwin, S. Ehrt et al. Science 2003 | sourish10 | IEP | 14671303 | Structural Analysis |
| Interaction | Regulatory Rv2111c | sourish10 | IEP | | Structural Analysis
KH. Darwin, S. Ehrt et al. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 2003 |
| Interaction | Regulatory Rv2097c | sourish10 | IEP | | Structural Analysis
KH. Darwin, S. Ehrt et al. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 2003 |
| Citation | SRS-A mediated bronchospasm by pharmacologic modification of lung anaphylaxis in vivo. authors,DM. Ritchie,JN. Sierchio,RJ. Capetola,ME. Rosenthale Agents Actions 1981 | sourish10 | IEP | 7282491 | Structural Analysis |
| Interaction | Regulatory Rv2111c | sourish10 | IEP | | Structural Analysis
authors,DM. Ritchie,JN. Sierchio,RJ. Capetola,ME. Rosenthale SRS-A mediated bronchospasm by pharmacologic modification of lung anaphylaxis in vivo. Agents Actions 1981 |
| Interaction | Regulatory Rv2097c | sourish10 | IEP | | Structural Analysis
authors,DM. Ritchie,JN. Sierchio,RJ. Capetola,ME. Rosenthale SRS-A mediated bronchospasm by pharmacologic modification of lung anaphylaxis in vivo. Agents Actions 1981 |
| Citation | SRS-A mediated bronchospasm by pharmacologic modification of lung anaphylaxis in vivo. authors,DM. Ritchie,JN. Sierchio,RJ. Capetola,ME. Rosenthale Agents Actions 1981 | sourish10 | IEP | 7282491 | Spectrophotometric Analysis |
| Interaction | Regulatory Rv2111c | sourish10 | IEP | | Spectrophotometric Analysis
authors,DM. Ritchie,JN. Sierchio,RJ. Capetola,ME. Rosenthale SRS-A mediated bronchospasm by pharmacologic modification of lung anaphylaxis in vivo. Agents Actions 1981 |
| Interaction | Regulatory Rv2097c | sourish10 | IEP | | Spectrophotometric Analysis
authors,DM. Ritchie,JN. Sierchio,RJ. Capetola,ME. Rosenthale SRS-A mediated bronchospasm by pharmacologic modification of lung anaphylaxis in vivo. Agents Actions 1981 |
| Citation | A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. M. Sutter,F. Striebel,FF. Damberger,FH. Allain,E. Weber-Ban FEBS Lett. 2009 | sourish10 | IEP | 19761766 | Structural Analysis |
| Interaction | Regulatory Rv2111c | sourish10 | IEP | | Structural Analysis
M. Sutter,F. Striebel,FF. Damberger,FH. Allain,E. Weber-Ban A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. FEBS Lett. 2009 |
| Interaction | Regulatory Rv2097c | sourish10 | IEP | | Structural Analysis
M. Sutter,F. Striebel,FF. Damberger,FH. Allain,E. Weber-Ban A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. FEBS Lett. 2009 |
| Citation | Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. KH. Darwin, G. Lin et al. Mol. Microbiol. 2005 | sourish10 | IEP | 15659170 | Structural Analysis |
| Interaction | Regulatory Rv2111c | sourish10 | IEP | | Structural Analysis
KH. Darwin, G. Lin et al. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol. Microbiol. 2005 |
| Interaction | Regulatory Rv2097c | sourish10 | IEP | | Structural Analysis
KH. Darwin, G. Lin et al. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol. Microbiol. 2005 |
| Citation | A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. M. Sutter,F. Striebel,FF. Damberger,FH. Allain,E. Weber-Ban FEBS Lett. 2009 | sourish10 | IEP | 19761766 | Spectrophotometric Analysis |
| Interaction | Regulatory Rv2111c | sourish10 | IEP | | Spectrophotometric Analysis
M. Sutter,F. Striebel,FF. Damberger,FH. Allain,E. Weber-Ban A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. FEBS Lett. 2009 |
| Interaction | Regulatory Rv2097c | sourish10 | IEP | | Spectrophotometric Analysis
M. Sutter,F. Striebel,FF. Damberger,FH. Allain,E. Weber-Ban A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. FEBS Lett. 2009 |
| Citation | Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. KH. Darwin, G. Lin et al. Mol. Microbiol. 2005 | sourish10 | IEP | 15659170 | Spectrophotometric Analysis |
| Interaction | Regulatory Rv2111c | sourish10 | IEP | | Spectrophotometric Analysis
KH. Darwin, G. Lin et al. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol. Microbiol. 2005 |
| Interaction | Regulatory Rv2097c | sourish10 | IEP | | Spectrophotometric Analysis
KH. Darwin, G. Lin et al. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol. Microbiol. 2005 |
| Citation | The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. KH. Darwin, S. Ehrt et al. Science 2003 | sourish10 | IEP | 14671303 | Spectrophotometric Analysis |
| Interaction | Regulatory Rv2111c | sourish10 | IEP | | Spectrophotometric Analysis
KH. Darwin, S. Ehrt et al. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 2003 |
| Interaction | Regulatory Rv2097c | sourish10 | IEP | | Spectrophotometric Analysis
KH. Darwin, S. Ehrt et al. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 2003 |
| Citation | Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. KH. Darwin, G. Lin et al. Mol. Microbiol. 2005 | ahal4789 | IEP | 15659170 | Structural Analysis |
| Interaction | Regulatory Rv2111c | ahal4789 | IEP | | Structural Analysis
KH. Darwin, G. Lin et al. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol. Microbiol. 2005 |
| Interaction | Regulatory Rv2097c | ahal4789 | IEP | | Structural Analysis
KH. Darwin, G. Lin et al. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol. Microbiol. 2005 |
| Citation | The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. KH. Darwin, S. Ehrt et al. Science 2003 | ahal4789 | IEP | 14671303 | Structural Analysis |
| Interaction | Regulatory Rv2111c | ahal4789 | IEP | | Structural Analysis
KH. Darwin, S. Ehrt et al. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 2003 |
| Interaction | Regulatory Rv2097c | ahal4789 | IEP | | Structural Analysis
KH. Darwin, S. Ehrt et al. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 2003 |
| Citation | SRS-A mediated bronchospasm by pharmacologic modification of lung anaphylaxis in vivo. authors,DM. Ritchie,JN. Sierchio,RJ. Capetola,ME. Rosenthale Agents Actions 1981 | ahal4789 | IEP | 7282491 | Structural Analysis |
| Interaction | Regulatory Rv2111c | ahal4789 | IEP | | Structural Analysis
authors,DM. Ritchie,JN. Sierchio,RJ. Capetola,ME. Rosenthale SRS-A mediated bronchospasm by pharmacologic modification of lung anaphylaxis in vivo. Agents Actions 1981 |
| Interaction | Regulatory Rv2097c | ahal4789 | IEP | | Structural Analysis
authors,DM. Ritchie,JN. Sierchio,RJ. Capetola,ME. Rosenthale SRS-A mediated bronchospasm by pharmacologic modification of lung anaphylaxis in vivo. Agents Actions 1981 |
| Citation | The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. KH. Darwin, S. Ehrt et al. Science 2003 | ahal4789 | IEP | 14671303 | Spectrophotometric Analysis |
| Interaction | Regulatory Rv2111c | ahal4789 | IEP | | Spectrophotometric Analysis
KH. Darwin, S. Ehrt et al. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 2003 |
| Interaction | Regulatory Rv2097c | ahal4789 | IEP | | Spectrophotometric Analysis
KH. Darwin, S. Ehrt et al. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 2003 |
| Citation | SRS-A mediated bronchospasm by pharmacologic modification of lung anaphylaxis in vivo. authors,DM. Ritchie,JN. Sierchio,RJ. Capetola,ME. Rosenthale Agents Actions 1981 | ahal4789 | IEP | 7282491 | Spectrophotometric Analysis |
| Interaction | Regulatory Rv2111c | ahal4789 | IEP | | Spectrophotometric Analysis
authors,DM. Ritchie,JN. Sierchio,RJ. Capetola,ME. Rosenthale SRS-A mediated bronchospasm by pharmacologic modification of lung anaphylaxis in vivo. Agents Actions 1981 |
| Interaction | Regulatory Rv2097c | ahal4789 | IEP | | Spectrophotometric Analysis
authors,DM. Ritchie,JN. Sierchio,RJ. Capetola,ME. Rosenthale SRS-A mediated bronchospasm by pharmacologic modification of lung anaphylaxis in vivo. Agents Actions 1981 |
| Citation | A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. M. Sutter,F. Striebel,FF. Damberger,FH. Allain,E. Weber-Ban FEBS Lett. 2009 | ahal4789 | IEP | 19761766 | Structural Analysis |
| Interaction | Regulatory Rv2111c | ahal4789 | IEP | | Structural Analysis
M. Sutter,F. Striebel,FF. Damberger,FH. Allain,E. Weber-Ban A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. FEBS Lett. 2009 |
| Interaction | Regulatory Rv2097c | ahal4789 | IEP | | Structural Analysis
M. Sutter,F. Striebel,FF. Damberger,FH. Allain,E. Weber-Ban A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. FEBS Lett. 2009 |
| Citation | A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. M. Sutter,F. Striebel,FF. Damberger,FH. Allain,E. Weber-Ban FEBS Lett. 2009 | ahal4789 | IEP | 19761766 | Spectrophotometric Analysis |
| Interaction | Regulatory Rv2111c | ahal4789 | IEP | | Spectrophotometric Analysis
M. Sutter,F. Striebel,FF. Damberger,FH. Allain,E. Weber-Ban A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. FEBS Lett. 2009 |
| Interaction | Regulatory Rv2097c | ahal4789 | IEP | | Spectrophotometric Analysis
M. Sutter,F. Striebel,FF. Damberger,FH. Allain,E. Weber-Ban A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. FEBS Lett. 2009 |
| Citation | Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. KH. Darwin, G. Lin et al. Mol. Microbiol. 2005 | ahal4789 | IEP | 15659170 | Spectrophotometric Analysis |
| Interaction | Regulatory Rv2111c | ahal4789 | IEP | | Spectrophotometric Analysis
KH. Darwin, G. Lin et al. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol. Microbiol. 2005 |
| Interaction | Regulatory Rv2097c | ahal4789 | IEP | | Spectrophotometric Analysis
KH. Darwin, G. Lin et al. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol. Microbiol. 2005 |