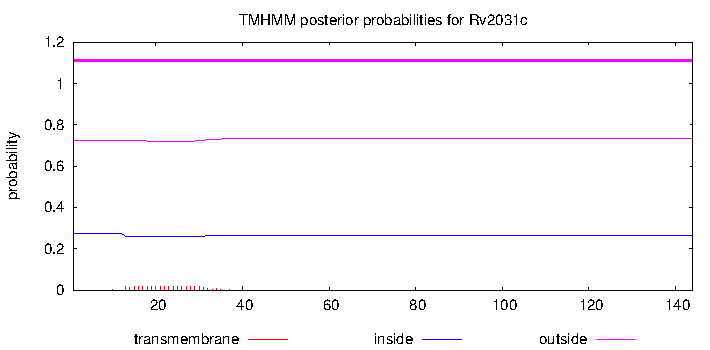
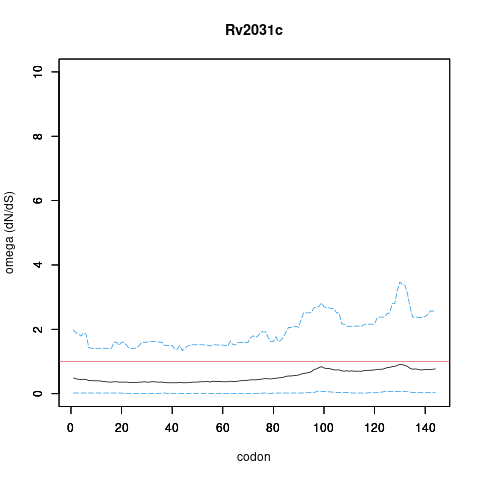
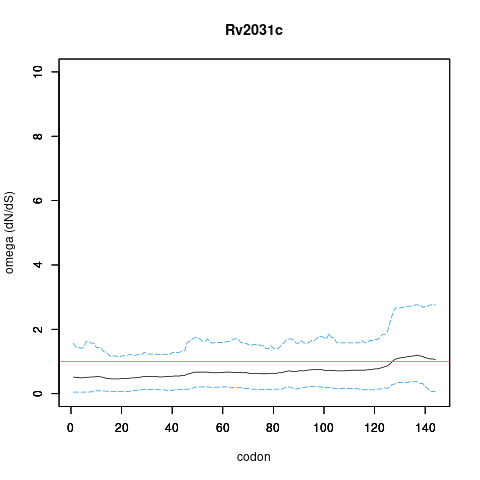
| Property | Value | Creator | Evidence | PMID | Comment |
| Interaction | Regulatory Rv2069 | ashwinigbhat | IEP | | Co-expression (Functional linkage)
R. Sun, PJ. Converse et al. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol. Microbiol. 2004 |
| Interaction | Regulatory Rv2069 | priti.priety | IEP | | Co-expression (Functional linkage)
R. Sun, PJ. Converse et al. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol. Microbiol. 2004 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | priti.priety | IDA | 19228590 | affinity purification |
| Interaction | Signaling Rv3133c | priti.priety | IDA | | affinity purification
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Recognition of stage-specific mycobacterial antigens differentiates between acute and latent infections with Mycobacterium tuberculosis. A. Demissie, EM. Leyten et al. Clin. Vaccine Immunol. 2006 | priti.priety | IDA | 16467323 | affinity purification |
| Interaction | Signaling Rv3133c | priti.priety | IDA | | affinity purification
A. Demissie, EM. Leyten et al. Recognition of stage-specific mycobacterial antigens differentiates between acute and latent infections with Mycobacterium tuberculosis. Clin. Vaccine Immunol. 2006 |
| Interaction | Regulatory Rv2032 | priti.priety | IDA | | Spectrophotometric analysis
A. Purkayastha, LA. McCue et al. Identification of a Mycobacterium tuberculosis putative classical nitroreductase gene whose expression is coregulated with that of the acr aene within macrophages, in standing versus shaking cultures, and under low oxygen conditions. Infect. Immun. 2002 |
| Interaction | Regulatory Rv2034 | priti.priety | IDA | | Bacterial one-hybrid system
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | Regulatory Rv2007c | akankshajain.21 | IEP | | co-expression(Functional linkage)
A. Kumar,JC. Toledo,RP. Patel,JR. Lancaster,AJ. Steyn Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. U.S.A. 2007 |
| Interaction | Regulatory Rv2007c | prabhakarsmail | IEP | | co-expression(Functional linkage)
A. Kumar,JC. Toledo,RP. Patel,JR. Lancaster,AJ. Steyn Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. U.S.A. 2007 |
| Interaction | Regulatory Rv1996 | priyadarshinipriyanka2001 | IEP | | Co-expression (Functional linkage)
MA. Florczyk, LA. McCue et al. A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires Rv3133c (dosR or devR) for expression. Infect. Immun. 2003 |
| Interaction | Regulatory Rv1996 | priyadarshinipriyanka2001 | IEP | | Co-expression (Functional linkage)
authors,SM. Hingley-Wilson,KE. Lougheed,K. Ferguson,S. Leiva,HD. Williams Individual Mycobacterium tuberculosis universal stress protein homologues are dispensable in vitro. Tuberculosis (Edinb) 2010 |
| Interaction | Regulatory Rv0757 | singhpankaj2116 | IEP | | Structural Analysis
J. Gonzalo-Asensio, S. Mostowy et al. PhoP: a missing piece in the intricate puzzle of Mycobacterium tuberculosis virulence. PLoS ONE 2008 |
| Interaction | Regulatory Rv0757 | singhpankaj2116 | IEP | | Spectrophotometric
J. Gonzalo-Asensio, S. Mostowy et al. PhoP: a missing piece in the intricate puzzle of Mycobacterium tuberculosis virulence. PLoS ONE 2008 |
| Interaction | RegulatedBy Rv0348 | yamir.moreno | IEP | | Microarrays. mRNA levels of regulated element measured and compared between wild-type and trans-element mutation (knockout, over expression etc.) performed by using microarray (or macroarray) experiments..
B. Abomoelak, EA. Hoye et al. mosR, a novel transcriptional regulator of hypoxia and virulence in Mycobacterium tuberculosis. J. Bacteriol. 2009 |
| Interaction | RegulatedBy Rv3416 | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | RegulatedBy Rv3678c | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | RegulatedBy Rv3681c | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | RegulatedBy Rv3133c | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | RegulatedBy Rv0445c | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | RegulatedBy Rv2034 | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | RegulatedBy Rv3133c | yamir.moreno | TAS | | Literature previously reported link (from Balazsi et al. 2008). Traceable author statement to experimental support.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Interaction | RegulatedBy Rv2069 | yamir.moreno | TAS | | Literature previously reported link (from Balazsi et al. 2008). Traceable author statement to experimental support.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |