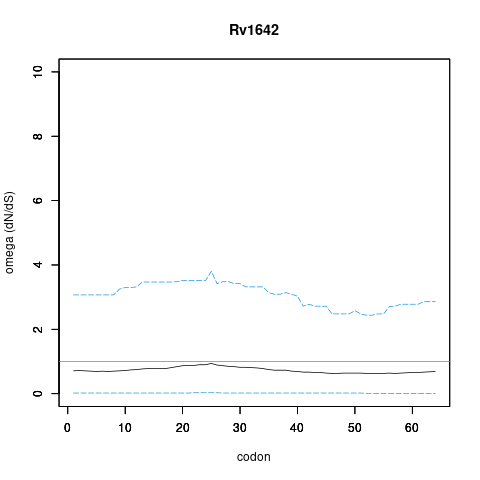
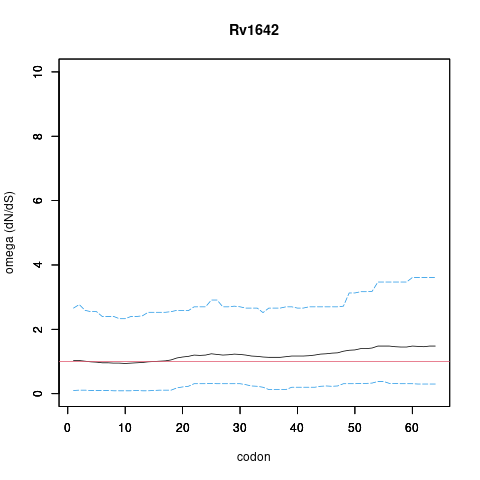
| Property | Value | Creator | Evidence | PMID | Comment |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Co-expression (Functional linkage)
authors,RJ. Smith,RG. Bryant Metal substitutions incarbonic anhydrase: a halide ion probe study. Biochem. Biophys. Res. Commun. 1975 |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Co-expression (Functional linkage)
authors,RJ. Smith,RG. Bryant Metal substitutions incarbonic anhydrase: a halide ion probe study. Biochem. Biophys. Res. Commun. 1975 |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Co-expression (Functional linkage)
authors,YW. Chow,R. Pietranico,A. Mukerji Studies of oxygen binding energy to hemoglobin molecule. Biochem. Biophys. Res. Commun. 1975 |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Co-expression (Functional linkage)
authors,F. Allemand,J. Haentjens,C. Chiaruttini,C. Royer,M. Springer Escherichia coli ribosomal protein L20 binds as a single monomer to its own mRNA bearing two potential binding sites. Nucleic Acids Res. 2007 |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Co-expression (Functional linkage)
authors,N. Choonee,S. Even,L. Zig,H. Putzer Ribosomal protein L20 controls expression of the Bacillus subtilis infC operon via a transcription attenuation mechanism. Nucleic Acids Res. 2007 |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Affinity purification (Physical interaction)
authors,YW. Chow,R. Pietranico,A. Mukerji Studies of oxygen binding energy to hemoglobin molecule. Biochem. Biophys. Res. Commun. 1975 |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Affinity purification (Physical interaction)
authors,RJ. Smith,RG. Bryant Metal substitutions incarbonic anhydrase: a halide ion probe study. Biochem. Biophys. Res. Commun. 1975 |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Affinity purification (Physical interaction)
authors,N. Choonee,S. Even,L. Zig,H. Putzer Ribosomal protein L20 controls expression of the Bacillus subtilis infC operon via a transcription attenuation mechanism. Nucleic Acids Res. 2007 |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Affinity purification (Physical interaction)
authors,RJ. Smith,RG. Bryant Metal substitutions incarbonic anhydrase: a halide ion probe study. Biochem. Biophys. Res. Commun. 1975 |
| Citation | Studies of oxygen binding energy to hemoglobin molecule. authors,YW. Chow,R. Pietranico,A. Mukerji Biochem. Biophys. Res. Commun. 1975 | sourish10 | ISO | 6 | Co-expression (Functional linkage) |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Co-expression (Functional linkage)
authors,YW. Chow,R. Pietranico,A. Mukerji Studies of oxygen binding energy to hemoglobin molecule. Biochem. Biophys. Res. Commun. 1975 |
| Citation | Metal substitutions incarbonic anhydrase: a halide ion probe study. authors,RJ. Smith,RG. Bryant Biochem. Biophys. Res. Commun. 1975 | sourish10 | ISO | 3 | Co-expression (Functional linkage) |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Co-expression (Functional linkage)
authors,RJ. Smith,RG. Bryant Metal substitutions incarbonic anhydrase: a halide ion probe study. Biochem. Biophys. Res. Commun. 1975 |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Affinity purification (Physical interaction)
authors,F. Allemand,J. Haentjens,C. Chiaruttini,C. Royer,M. Springer Escherichia coli ribosomal protein L20 binds as a single monomer to its own mRNA bearing two potential binding sites. Nucleic Acids Res. 2007 |
| Citation | Studies of oxygen binding energy to hemoglobin molecule. authors,YW. Chow,R. Pietranico,A. Mukerji Biochem. Biophys. Res. Commun. 1975 | sourish10 | ISO | 6 | Affinity purification (Physical interaction) |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Affinity purification (Physical interaction)
authors,YW. Chow,R. Pietranico,A. Mukerji Studies of oxygen binding energy to hemoglobin molecule. Biochem. Biophys. Res. Commun. 1975 |
| Citation | Metal substitutions incarbonic anhydrase: a halide ion probe study. authors,RJ. Smith,RG. Bryant Biochem. Biophys. Res. Commun. 1975 | sourish10 | ISO | 3 | Affinity purification (Physical interaction) |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Affinity purification (Physical interaction)
authors,RJ. Smith,RG. Bryant Metal substitutions incarbonic anhydrase: a halide ion probe study. Biochem. Biophys. Res. Commun. 1975 |
| Citation | Escherichia coli ribosomal protein L20 binds as a single monomer to its own mRNA bearing two potential binding sites. authors,F. Allemand,J. Haentjens,C. Chiaruttini,C. Royer,M. Springer Nucleic Acids Res. 2007 | sourish10 | ISO | 17439971 | Co-expression (Functional linkage) |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Co-expression (Functional linkage)
authors,F. Allemand,J. Haentjens,C. Chiaruttini,C. Royer,M. Springer Escherichia coli ribosomal protein L20 binds as a single monomer to its own mRNA bearing two potential binding sites. Nucleic Acids Res. 2007 |
| Citation | Ribosomal protein L20 controls expression of the Bacillus subtilis infC operon via a transcription attenuation mechanism. authors,N. Choonee,S. Even,L. Zig,H. Putzer Nucleic Acids Res. 2007 | sourish10 | ISO | 17289755 | Co-expression (Functional linkage) |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Co-expression (Functional linkage)
authors,N. Choonee,S. Even,L. Zig,H. Putzer Ribosomal protein L20 controls expression of the Bacillus subtilis infC operon via a transcription attenuation mechanism. Nucleic Acids Res. 2007 |
| Citation | Metal substitutions incarbonic anhydrase: a halide ion probe study. authors,RJ. Smith,RG. Bryant Biochem. Biophys. Res. Commun. 1975 | sourish10 | ISO | 3 | Co-expression (Functional linkage) |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Co-expression (Functional linkage)
authors,RJ. Smith,RG. Bryant Metal substitutions incarbonic anhydrase: a halide ion probe study. Biochem. Biophys. Res. Commun. 1975 |
| Citation | Escherichia coli ribosomal protein L20 binds as a single monomer to its own mRNA bearing two potential binding sites. authors,F. Allemand,J. Haentjens,C. Chiaruttini,C. Royer,M. Springer Nucleic Acids Res. 2007 | sourish10 | ISO | 17439971 | Affinity purification (Physical interaction) |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Affinity purification (Physical interaction)
authors,F. Allemand,J. Haentjens,C. Chiaruttini,C. Royer,M. Springer Escherichia coli ribosomal protein L20 binds as a single monomer to its own mRNA bearing two potential binding sites. Nucleic Acids Res. 2007 |
| Citation | Ribosomal protein L20 controls expression of the Bacillus subtilis infC operon via a transcription attenuation mechanism. authors,N. Choonee,S. Even,L. Zig,H. Putzer Nucleic Acids Res. 2007 | sourish10 | ISO | 17289755 | Affinity purification (Physical interaction) |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Affinity purification (Physical interaction)
authors,N. Choonee,S. Even,L. Zig,H. Putzer Ribosomal protein L20 controls expression of the Bacillus subtilis infC operon via a transcription attenuation mechanism. Nucleic Acids Res. 2007 |
| Citation | Metal substitutions incarbonic anhydrase: a halide ion probe study. authors,RJ. Smith,RG. Bryant Biochem. Biophys. Res. Commun. 1975 | sourish10 | ISO | 3 | Affinity purification (Physical interaction) |
| Interaction | Transcription Rv1643 | sourish10 | ISO | | Affinity purification (Physical interaction)
authors,RJ. Smith,RG. Bryant Metal substitutions incarbonic anhydrase: a halide ion probe study. Biochem. Biophys. Res. Commun. 1975 |