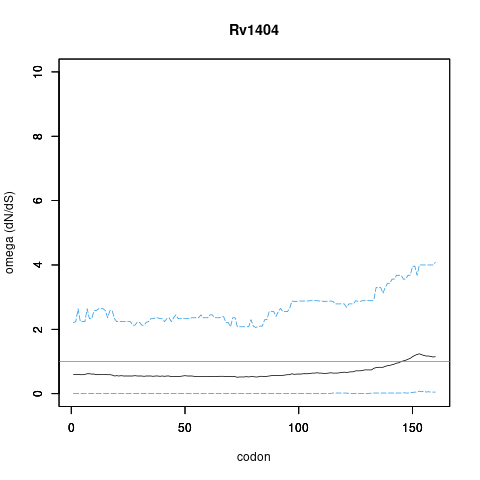
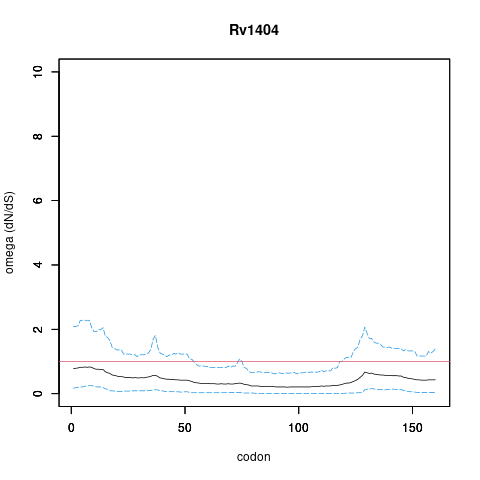
| Property | Value | Creator | Evidence | PMID | Comment |
| Interaction | Regulatory Rv3532 | raj252000 | IEP | | Co-expression (Functional linkage)
P. Golby, J. Nunez et al. Characterization of two in vivo-expressed methyltransferases of the Mycobacterium tuberculosis complex: antigenicity and genetic regulation. Microbiology (Reading, Engl.) 2008 |
| Interaction | Regulatory Rv3533c | salluamity1 | IEP | | Co-expression (Functional linkage)
P. Golby, J. Nunez et al. Characterization of two in vivo-expressed methyltransferases of the Mycobacterium tuberculosis complex: antigenicity and genetic regulation. Microbiology (Reading, Engl.) 2008 |
| Interaction | Regulatory Rv3533c | salluamity1 | IEP | | Co-expression (Functional linkage)
MI. Voskuil,D. Schnappinger,R. Rutherford,Y. Liu,GK. Schoolnik Regulation of the Mycobacterium tuberculosis PE/PPE genes. Tuberculosis (Edinb) 2004 |
| Interaction | Regulatory Rv3531c | raj252000 | IEP | | Co-expression (Functional linkage)
SL. Kendall, M. Withers et al. A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis. Mol. Microbiol. 2007 |
| Interaction | Regulatory Rv3531c | raj252000 | IEP | | Co-expression (Functional linkage)
P. Golby, J. Nunez et al. Characterization of two in vivo-expressed methyltransferases of the Mycobacterium tuberculosis complex: antigenicity and genetic regulation. Microbiology (Reading, Engl.) 2008 |
| Interaction | PhysicalInteraction Rv1931c | swatigandhi19 | IMP | | Co-expression (Functional Linkage)
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Interaction | PhysicalInteraction Rv1931c | swatigandhi19 | IMP | | Co-expression (Functional Linkage)
CC. Frota, KG. Papavinasasundaram et al. The AraC family transcriptional regulator Rv1931c plays a role in the virulence of Mycobacterium tuberculosis. Infect. Immun. 2004 |
| Interaction | Regulatory Rv1403c | ashwinigbhat | IEP | | qRT-PCR
P. Golby, J. Nunez et al. Characterization of two in vivo-expressed methyltransferases of the Mycobacterium tuberculosis complex: antigenicity and genetic regulation. Microbiology (Reading, Engl.) 2008 |
| Interaction | Regulatory Rv1405c | ashwinigbhat | IEP | | qRT-PCR
P. Golby, J. Nunez et al. Characterization of two in vivo-expressed methyltransferases of the Mycobacterium tuberculosis complex: antigenicity and genetic regulation. Microbiology (Reading, Engl.) 2008 |
| Citation | IdeR in mycobacteria: from target recognition to physiological function. S. Ranjan, S. Yellaboina et al. Crit. Rev. Microbiol. 2006 | ashwinigbhat | TAS | 16809230 | positional relative entropy method |
| Interaction | Regulatory Rv2711 | ashwinigbhat | TAS | | positional relative entropy method
S. Ranjan, S. Yellaboina et al. IdeR in mycobacteria: from target recognition to physiological function. Crit. Rev. Microbiol. 2006 |
| Citation | Computational prediction and experimental verification of novel IdeR binding sites in the upstream sequences of Mycobacterium tuberculosis open reading frames. P. Prakash, S. Yellaboina et al. Bioinformatics 2005 | ashwinigbhat | TAS | 15746274 | positional relative entropy method |
| Interaction | Regulatory Rv2711 | ashwinigbhat | TAS | | positional relative entropy method
P. Prakash, S. Yellaboina et al. Computational prediction and experimental verification of novel IdeR binding sites in the upstream sequences of Mycobacterium tuberculosis open reading frames. Bioinformatics 2005 |
| Citation | The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. G. Balzsi, AP. Heath et al. Mol. Syst. Biol. 2008 | ashwinigbhat | IEP | 18985025 | Microarray analysis |
| Interaction | Regulatory Rv1931c | ashwinigbhat | IEP | | Microarray analysis
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Citation | Characterization of two in vivo-expressed methyltransferases of the Mycobacterium tuberculosis complex: antigenicity and genetic regulation. P. Golby, J. Nunez et al. Microbiology (Reading, Engl.) 2008 | ashwinigbhat | IEP | 18375799 | qRT-PCR |
| Interaction | Transcription Rv0193c | sourish10 | IMP | | Protein-Protein
P. Golby, J. Nunez et al. Characterization of two in vivo-expressed methyltransferases of the Mycobacterium tuberculosis complex: antigenicity and genetic regulation. Microbiology (Reading, Engl.) 2008 |
| Interaction | Regulatory Rv0195 | aparna.vchalam | IEP | | Co-expression (Functional linkage)
P. Golby, J. Nunez et al. Characterization of two in vivo-expressed methyltransferases of the Mycobacterium tuberculosis complex: antigenicity and genetic regulation. Microbiology (Reading, Engl.) 2008 |
| Interaction | RegulatedBy Rv1931c | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Citation | The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. G. Balzsi, AP. Heath et al. Mol. Syst. Biol. 2008 | yamir.moreno | ISO | 18985025 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Interaction | Regulates Rv1404 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Interaction | RegulatedBy Rv1404 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Citation | The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. G. Balzsi, AP. Heath et al. Mol. Syst. Biol. 2008 | yamir.moreno | ISO | 18985025 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Interaction | Regulates Rv1931c | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Interaction | RegulatedBy Rv1931c | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
authors,M. Madan Babu,SA. Teichmann,L. Aravind Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006 |
| Citation | Evolutionary dynamics of prokaryotic transcriptional regulatory networks. authors,M. Madan Babu,SA. Teichmann,L. Aravind J. Mol. Biol. 2006 | yamir.moreno | ISO | 16530225 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Interaction | Regulates Rv1404 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
authors,M. Madan Babu,SA. Teichmann,L. Aravind Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006 |
| Interaction | RegulatedBy Rv1404 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
authors,M. Madan Babu,SA. Teichmann,L. Aravind Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006 |
| Citation | Evolutionary dynamics of prokaryotic transcriptional regulatory networks. authors,M. Madan Babu,SA. Teichmann,L. Aravind J. Mol. Biol. 2006 | yamir.moreno | ISO | 16530225 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Interaction | Regulates Rv1931c | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
authors,M. Madan Babu,SA. Teichmann,L. Aravind Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006 |