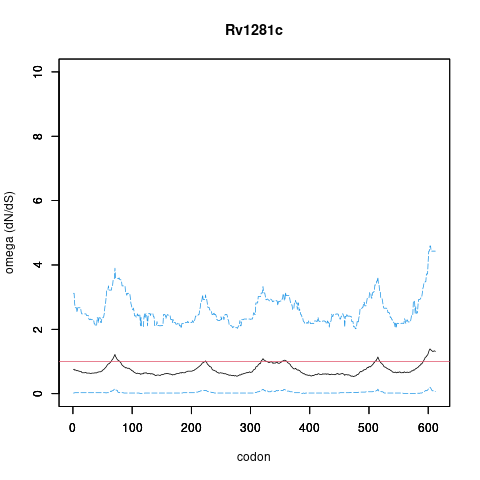
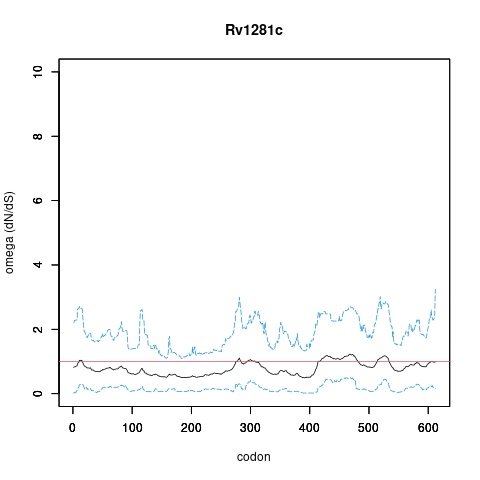
| Property | Value | Creator | Evidence | PMID | Comment |
| Interaction | PhysicalInteraction Rv1283c | swetha.r | IGC | | Operon (Functional linkage)
MA. Flores-Valdez,RP. Morris,F. Laval,M. Daff,GK. Schoolnik Mycobacterium tuberculosis modulates its cell surface via an oligopeptide permease (Opp) transport system. FASEB J. 2009 |
| Interaction | PhysicalInteraction Rv1283c | swetha.r | IGC | | Operon (Functional linkage)
authors,M. Braibant,P. Gilot,J. Content The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2000 |
| Interaction | PhysicalInteraction Rv1283c | swetha.r | IGC | | Operon (Functional linkage)
authors,YK. Dayaram,MT. Talaue,ND. Connell,V. Venketaraman Characterization of a glutathione metabolic mutant of Mycobacterium tuberculosis and its resistance to glutathione and nitrosoglutathione. J. Bacteriol. 2006 |
| Interaction | PhysicalInteraction Rv1282c | swetha.r | IGC | | Operon (Functional linkage)
MA. Flores-Valdez,RP. Morris,F. Laval,M. Daff,GK. Schoolnik Mycobacterium tuberculosis modulates its cell surface via an oligopeptide permease (Opp) transport system. FASEB J. 2009 |
| Interaction | PhysicalInteraction Rv1283c | swetha.r | IGC | | Operon (Functional linkage)
RM. Green, A. Seth et al. A peptide permease mutant of Mycobacterium bovis BCG resistant to the toxic peptides glutathione and S-nitrosoglutathione. Infect. Immun. 2000 |
| Interaction | PhysicalInteraction Rv1282c | swetha.r | IGC | | Operon (Functional linkage)
authors,M. Braibant,P. Gilot,J. Content The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2000 |
| Interaction | PhysicalInteraction Rv1282c | swetha.r | IGC | | Operon (Functional linkage)
authors,YK. Dayaram,MT. Talaue,ND. Connell,V. Venketaraman Characterization of a glutathione metabolic mutant of Mycobacterium tuberculosis and its resistance to glutathione and nitrosoglutathione. J. Bacteriol. 2006 |
| Interaction | PhysicalInteraction Rv1283c | swetha.r | IGC | | Operon (Functional linkage)
MA. Flores-Valdez,RP. Morris,F. Laval,M. Daff,GK. Schoolnik Mycobacterium tuberculosis modulates its cell surface via an oligopeptide permease (Opp) transport system. FASEB J. 2009 |
| Interaction | PhysicalInteraction Rv1282c | swetha.r | IGC | | Operon (Functional linkage)
MA. Flores-Valdez,RP. Morris,F. Laval,M. Daff,GK. Schoolnik Mycobacterium tuberculosis modulates its cell surface via an oligopeptide permease (Opp) transport system. FASEB J. 2009 |
| Interaction | PhysicalInteraction Rv1280c | swetha.r | IGC | | Operon (Functional linkage)
MA. Flores-Valdez,RP. Morris,F. Laval,M. Daff,GK. Schoolnik Mycobacterium tuberculosis modulates its cell surface via an oligopeptide permease (Opp) transport system. FASEB J. 2009 |
| Interaction | PhysicalInteraction Rv1282c | swetha.r | IGC | | Operon (Functional linkage)
RM. Green, A. Seth et al. A peptide permease mutant of Mycobacterium bovis BCG resistant to the toxic peptides glutathione and S-nitrosoglutathione. Infect. Immun. 2000 |
| Interaction | PhysicalInteraction Rv1283c | swetha.r | IGC | | Operon (Functional linkage)
authors,M. Braibant,P. Gilot,J. Content The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2000 |
| Interaction | PhysicalInteraction Rv1282c | swetha.r | IGC | | Operon (Functional linkage)
authors,M. Braibant,P. Gilot,J. Content The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2000 |
| Interaction | PhysicalInteraction Rv1280c | swetha.r | IGC | | Operon (Functional linkage)
authors,M. Braibant,P. Gilot,J. Content The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2000 |
| Citation | Characterization of a glutathione metabolic mutant of Mycobacterium tuberculosis and its resistance to glutathione and nitrosoglutathione. authors,YK. Dayaram,MT. Talaue,ND. Connell,V. Venketaraman J. Bacteriol. 2006 | swetha.r | IGC | 16452418 | Operon (Functional linkage) |
| Interaction | PhysicalInteraction Rv1283c | swetha.r | IGC | | Operon (Functional linkage)
authors,YK. Dayaram,MT. Talaue,ND. Connell,V. Venketaraman Characterization of a glutathione metabolic mutant of Mycobacterium tuberculosis and its resistance to glutathione and nitrosoglutathione. J. Bacteriol. 2006 |
| Interaction | PhysicalInteraction Rv1282c | swetha.r | IGC | | Operon (Functional linkage)
authors,YK. Dayaram,MT. Talaue,ND. Connell,V. Venketaraman Characterization of a glutathione metabolic mutant of Mycobacterium tuberculosis and its resistance to glutathione and nitrosoglutathione. J. Bacteriol. 2006 |
| Interaction | PhysicalInteraction Rv1280c | swetha.r | IGC | | Operon (Functional linkage)
authors,YK. Dayaram,MT. Talaue,ND. Connell,V. Venketaraman Characterization of a glutathione metabolic mutant of Mycobacterium tuberculosis and its resistance to glutathione and nitrosoglutathione. J. Bacteriol. 2006 |
| Citation | Mycobacterium tuberculosis modulates its cell surface via an oligopeptide permease (Opp) transport system. MA. Flores-Valdez,RP. Morris,F. Laval,M. Daff,GK. Schoolnik FASEB J. 2009 | swetha.r | IGC | 19671666 | Operon (Functional linkage) |
| Interaction | PhysicalInteraction Rv1280c | swetha.r | IGC | | Operon (Functional linkage)
MA. Flores-Valdez,RP. Morris,F. Laval,M. Daff,GK. Schoolnik Mycobacterium tuberculosis modulates its cell surface via an oligopeptide permease (Opp) transport system. FASEB J. 2009 |
| Citation | A peptide permease mutant of Mycobacterium bovis BCG resistant to the toxic peptides glutathione and S-nitrosoglutathione. RM. Green, A. Seth et al. Infect. Immun. 2000 | swetha.r | IGC | 10639400 | Operon (Functional linkage) |
| Interaction | PhysicalInteraction Rv1283c | swetha.r | IGC | | Operon (Functional linkage)
RM. Green, A. Seth et al. A peptide permease mutant of Mycobacterium bovis BCG resistant to the toxic peptides glutathione and S-nitrosoglutathione. Infect. Immun. 2000 |
| Interaction | PhysicalInteraction Rv1282c | swetha.r | IGC | | Operon (Functional linkage)
RM. Green, A. Seth et al. A peptide permease mutant of Mycobacterium bovis BCG resistant to the toxic peptides glutathione and S-nitrosoglutathione. Infect. Immun. 2000 |
| Interaction | PhysicalInteraction Rv1280c | swetha.r | IGC | | Operon (Functional linkage)
RM. Green, A. Seth et al. A peptide permease mutant of Mycobacterium bovis BCG resistant to the toxic peptides glutathione and S-nitrosoglutathione. Infect. Immun. 2000 |
| Citation | The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. authors,M. Braibant,P. Gilot,J. Content FEMS Microbiol. Rev. 2000 | swetha.r | IGC | 10978546 | Operon (Functional linkage) |
| Interaction | PhysicalInteraction Rv1280c | swetha.r | IGC | | Operon (Functional linkage)
authors,M. Braibant,P. Gilot,J. Content The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2000 |
| Interaction | PhysicalInteraction Rv1280c | swetha.r | IGC | | Operon (Functional linkage)
authors,YK. Dayaram,MT. Talaue,ND. Connell,V. Venketaraman Characterization of a glutathione metabolic mutant of Mycobacterium tuberculosis and its resistance to glutathione and nitrosoglutathione. J. Bacteriol. 2006 |
| Interaction | PhysicalInteraction Rv1280c | swetha.r | IGC | | Operon (Functional linkage)
RM. Green, A. Seth et al. A peptide permease mutant of Mycobacterium bovis BCG resistant to the toxic peptides glutathione and S-nitrosoglutathione. Infect. Immun. 2000 |
| Interaction | RegulatedBy Rv3291c | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Interaction | RegulatedBy Rv0491 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Interaction | RegulatedBy Rv3286c | yamir.moreno | IEP | | Microarrays. mRNA levels of regulated element measured and compared between wild-type and trans-element mutation (knockout, over expression etc.) performed by using microarray (or macroarray) experiments..
EP. Williams, JH. Lee et al. Mycobacterium tuberculosis SigF regulates genes encoding cell wall-associated proteins and directly regulates the transcriptional regulatory gene phoY1. J. Bacteriol. 2007 |
| Interaction | RegulatedBy Rv3291c | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
authors,M. Madan Babu,SA. Teichmann,L. Aravind Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006 |