| Property | Value | Creator | Evidence | PMID | Comment |
| Interaction | Signaling Rv3330 | hibeeluck | IDA | | Spectrophotometric
X. Zheng, KG. Papavinasasundaram et al. Novel substrates of Mycobacterium tuberculosis PknH Ser/Thr kinase. Biochem. Biophys. Res. Commun. 2007 |
| Interaction | Signaling Rv3330 | hibeeluck | IDA | | Structural Analysis
X. Zheng, KG. Papavinasasundaram et al. Novel substrates of Mycobacterium tuberculosis PknH Ser/Thr kinase. Biochem. Biophys. Res. Commun. 2007 |
| Interaction | Signaling Rv3330 | jhum4u2006 | IDA | | Spectrophotometric
X. Zheng, KG. Papavinasasundaram et al. Novel substrates of Mycobacterium tuberculosis PknH Ser/Thr kinase. Biochem. Biophys. Res. Commun. 2007 |
| Interaction | Signaling Rv3330 | jhum4u2006 | IDA | | Structural Analysis
X. Zheng, KG. Papavinasasundaram et al. Novel substrates of Mycobacterium tuberculosis PknH Ser/Thr kinase. Biochem. Biophys. Res. Commun. 2007 |
| Interaction | Signaling Rv2133c | akankshajain.21 | NAS | | Co-occurance (Functional Linkage)
authors,MA. Krasilnikov Phosphatidylinositol-3 kinase dependent pathways: the role in control of cell growth, survival, and malignant transformation. Biochemistry Mosc. 2000 |
| Interaction | Regulatory Rv3793 | shahanup86 | NAS | | Affinity purification (Physical interaction)
K. Sharma, M. Gupta et al. Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. J. Bacteriol. 2006 |
| Interaction | Signaling Rv1267c | shruti4rana | IDA | | Structural Analysis
LJ. Alderwick, V. Molle et al. Molecular structure of EmbR, a response element of Ser/Thr kinase signaling in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 2006 |
| Interaction | Signaling Rv1267c | shruti4rana | IDA | | Structural Analysis
K. Sharma, M. Gupta et al. Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. J. Bacteriol. 2006 |
| Citation | Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. K. Sharma, M. Gupta et al. J. Bacteriol. 2006 | shahanup86 | IPI | 16585755 | Affinity purification (Physical interaction) |
| Interaction | Activation Rv1267c | shahanup86 | IPI | | Affinity purification (Physical interaction)
K. Sharma, M. Gupta et al. Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. J. Bacteriol. 2006 |
| Citation | Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. K. Sharma, M. Gupta et al. J. Bacteriol. 2006 | shahanup86 | NAS | 16585755 | Affinity purification (Physical interaction) |
| Interaction | Regulatory Rv3794 | shahanup86 | NAS | | Affinity purification (Physical interaction)
K. Sharma, M. Gupta et al. Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. J. Bacteriol. 2006 |
| Interaction | Regulatory Rv3795 | shahanup86 | NAS | | Affinity purification (Physical interaction)
K. Sharma, M. Gupta et al. Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. J. Bacteriol. 2006 |
| Interaction | Signaling Rv0681 | sudhakar_dipu | IDA | | Spectrophotometric
X. Zheng, KG. Papavinasasundaram et al. Novel substrates of Mycobacterium tuberculosis PknH Ser/Thr kinase. Biochem. Biophys. Res. Commun. 2007 |
| Interaction | Signaling Rv0681 | sudhakar_dipu | IDA | | Structural Analysis
X. Zheng, KG. Papavinasasundaram et al. Novel substrates of Mycobacterium tuberculosis PknH Ser/Thr kinase. Biochem. Biophys. Res. Commun. 2007 |
| Citation | Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. K. Sharma, M. Gupta et al. J. Bacteriol. 2006 | yamir.moreno | IEP | 16585755 | qRT-PCR. mRNA expression levels of regulated element measured and compared between wild-type and trans-element mutation (knockout, over expression etc.) performed by using qRT-PCR technique. |
| Interaction | Regulates Rv1267c | yamir.moreno | IEP | | qRT-PCR. mRNA expression levels of regulated element measured and compared between wild-type and trans-element mutation (knockout, over expression etc.) performed by using qRT-PCR technique.
K. Sharma, M. Gupta et al. Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. J. Bacteriol. 2006 |
| Citation | The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. G. Balzsi, AP. Heath et al. Mol. Syst. Biol. 2008 | yamir.moreno | TAS | 18985025 | Literature previously reported link (from Balazsi et al. 2008). Traceable author statement to experimental support. |
| Interaction | Regulates Rv1267c | yamir.moreno | TAS | | Literature previously reported link (from Balazsi et al. 2008). Traceable author statement to experimental support.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
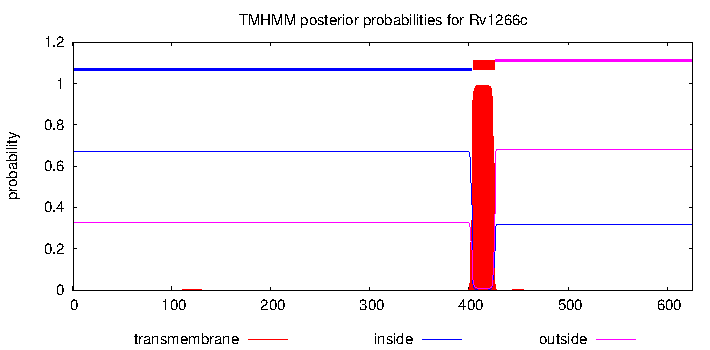
| Name | TRANSMEMBRANE SERINE/THREONINE-PROTEIN KINASE, known to phosphorylate InhA;KasA, KasB | mjackson | IDA | | Regulatory proteins |