| Property | Value | Creator | Evidence | PMID | Comment |
| Interaction | Regulatory Rv2939 | ashwinigbhat | IDA | | One hybrid System
DM. Collins, B. Skou et al. Generation of attenuated Mycobacterium bovis strains by signature-tagged mutagenesis for discovery of novel vaccine candidates. Infect. Immun. 2005 |
| Interaction | Regulatory Rv2939 | ashwinigbhat | IDA | | One hybrid System
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | Regulatory Rv2935 | ashwinigbhat | IDA | | One hybrid System
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | Regulatory Rv2935 | ashwinigbhat | IDA | | One hybrid System
DM. Collins, B. Skou et al. Generation of attenuated Mycobacterium bovis strains by signature-tagged mutagenesis for discovery of novel vaccine candidates. Infect. Immun. 2005 |
| Interaction | Regulatory Rv2934 | ashwinigbhat | IDA | | One hybrid System
DM. Collins, B. Skou et al. Generation of attenuated Mycobacterium bovis strains by signature-tagged mutagenesis for discovery of novel vaccine candidates. Infect. Immun. 2005 |
| Interaction | Regulatory Rv2934 | ashwinigbhat | IDA | | One hybrid System
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | Regulatory Rv2933 | ashwinigbhat | IDA | | One hybrid System
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | Regulatory Rv2933 | ashwinigbhat | IDA | | One hybrid System
DM. Collins, B. Skou et al. Generation of attenuated Mycobacterium bovis strains by signature-tagged mutagenesis for discovery of novel vaccine candidates. Infect. Immun. 2005 |
| Interaction | Regulatory Rv2932 | ashwinigbhat | IDA | | One hybrid System
DM. Collins, B. Skou et al. Generation of attenuated Mycobacterium bovis strains by signature-tagged mutagenesis for discovery of novel vaccine candidates. Infect. Immun. 2005 |
| Interaction | Regulatory Rv2932 | ashwinigbhat | IDA | | One hybrid System
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | Regulatory Rv2931 | ashwinigbhat | IDA | | One hybrid System
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | Regulatory Rv2931 | ashwinigbhat | IDA | | One hybrid System
DM. Collins, B. Skou et al. Generation of attenuated Mycobacterium bovis strains by signature-tagged mutagenesis for discovery of novel vaccine candidates. Infect. Immun. 2005 |
| Interaction | Regulatory Rv2930 | ashwinigbhat | IDA | | One hybrid System
DM. Collins, B. Skou et al. Generation of attenuated Mycobacterium bovis strains by signature-tagged mutagenesis for discovery of novel vaccine candidates. Infect. Immun. 2005 |
| Interaction | Regulatory Rv2930 | ashwinigbhat | IDA | | One hybrid System
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | Regulatory Rv1911c | shahanup86 | IEP | | Co-expression (Functional linkage)
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | Regulatory Rv1912c | shahanup86 | IEP | | Co-expression (Functional linkage)
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | shahanup86 | IEP | 19228590 | Co-expression (Functional linkage) |
| Interaction | Regulatory Rv1907c | shahanup86 | IEP | | Co-expression (Functional linkage)
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | Regulatory Rv1908c | shahanup86 | IEP | | Co-expression (Functional linkage)
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | Regulatory Rv1909c | shahanup86 | IEP | | Co-expression (Functional linkage)
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | Regulatory Rv1910c | shahanup86 | IEP | | Co-expression (Functional linkage)
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Interaction | RegulatedBy Rv0182c | yamir.moreno | IEP | | Microarrays. mRNA levels of regulated element measured and compared between wild-type and trans-element mutation (knockout, over expression etc.) performed by using microarray (or macroarray) experiments..
JH. Lee, DE. Geiman et al. Role of stress response sigma factor SigG in Mycobacterium tuberculosis. J. Bacteriol. 2008 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv2938 | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv2939 | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv3537 | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv3538 | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv2934 | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv2935 | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv2936 | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv2937 | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv2930 | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv2931 | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv2932 | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv2933 | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv1909c | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv1910c | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv1911c | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv1912c | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv1907c | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. M. Guo, H. Feng et al. Genome Res. 2009 | yamir.moreno | IDA | 19228590 | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. . |
| Interaction | Regulates Rv1908c | yamir.moreno | IDA | | One hybrid reporter system. Physical binding of the regulator to the regulated promoter proved by using electrophoretic mobility shift assay. .
M. Guo, H. Feng et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009 |
| Citation | The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. G. Balzsi, AP. Heath et al. Mol. Syst. Biol. 2008 | yamir.moreno | ISO | 18985025 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Interaction | Regulates Rv2428 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Citation | The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. G. Balzsi, AP. Heath et al. Mol. Syst. Biol. 2008 | yamir.moreno | ISO | 18985025 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Interaction | Regulates Rv3913 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Citation | The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. G. Balzsi, AP. Heath et al. Mol. Syst. Biol. 2008 | yamir.moreno | ISO | 18985025 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
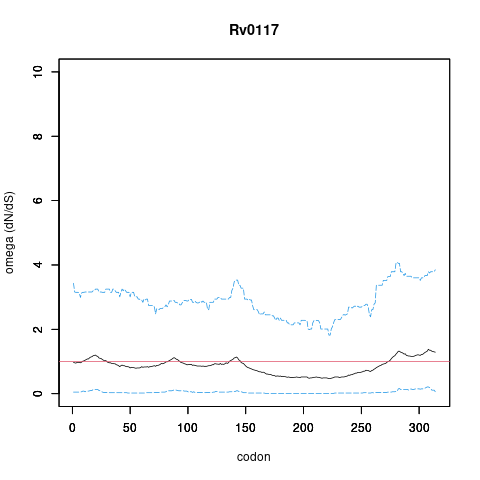
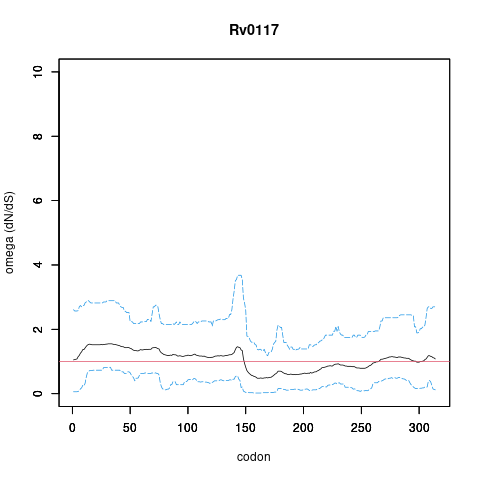
| Interaction | Regulates Rv0117 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Interaction | RegulatedBy Rv0117 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Citation | The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. G. Balzsi, AP. Heath et al. Mol. Syst. Biol. 2008 | yamir.moreno | ISO | 18985025 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Interaction | Regulates Rv1908c | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
G. Balzsi, AP. Heath et al. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008 |
| Interaction | Regulates Rv3913 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
authors,M. Madan Babu,SA. Teichmann,L. Aravind Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006 |
| Citation | Evolutionary dynamics of prokaryotic transcriptional regulatory networks. authors,M. Madan Babu,SA. Teichmann,L. Aravind J. Mol. Biol. 2006 | yamir.moreno | ISO | 16530225 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Interaction | Regulates Rv0117 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
authors,M. Madan Babu,SA. Teichmann,L. Aravind Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006 |
| Interaction | RegulatedBy Rv0117 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
authors,M. Madan Babu,SA. Teichmann,L. Aravind Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006 |
| Citation | Evolutionary dynamics of prokaryotic transcriptional regulatory networks. authors,M. Madan Babu,SA. Teichmann,L. Aravind J. Mol. Biol. 2006 | yamir.moreno | ISO | 16530225 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Interaction | Regulates Rv1908c | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
authors,M. Madan Babu,SA. Teichmann,L. Aravind Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006 |
| Citation | Evolutionary dynamics of prokaryotic transcriptional regulatory networks. authors,M. Madan Babu,SA. Teichmann,L. Aravind J. Mol. Biol. 2006 | yamir.moreno | ISO | 16530225 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |
| Interaction | Regulates Rv2428 | yamir.moreno | ISO | | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli.
authors,M. Madan Babu,SA. Teichmann,L. Aravind Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006 |
| Citation | Evolutionary dynamics of prokaryotic transcriptional regulatory networks. authors,M. Madan Babu,SA. Teichmann,L. Aravind J. Mol. Biol. 2006 | yamir.moreno | ISO | 16530225 | E.coli orthology based inference. Orthologous pair regulator-target found in E.coli. |